欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/135702185
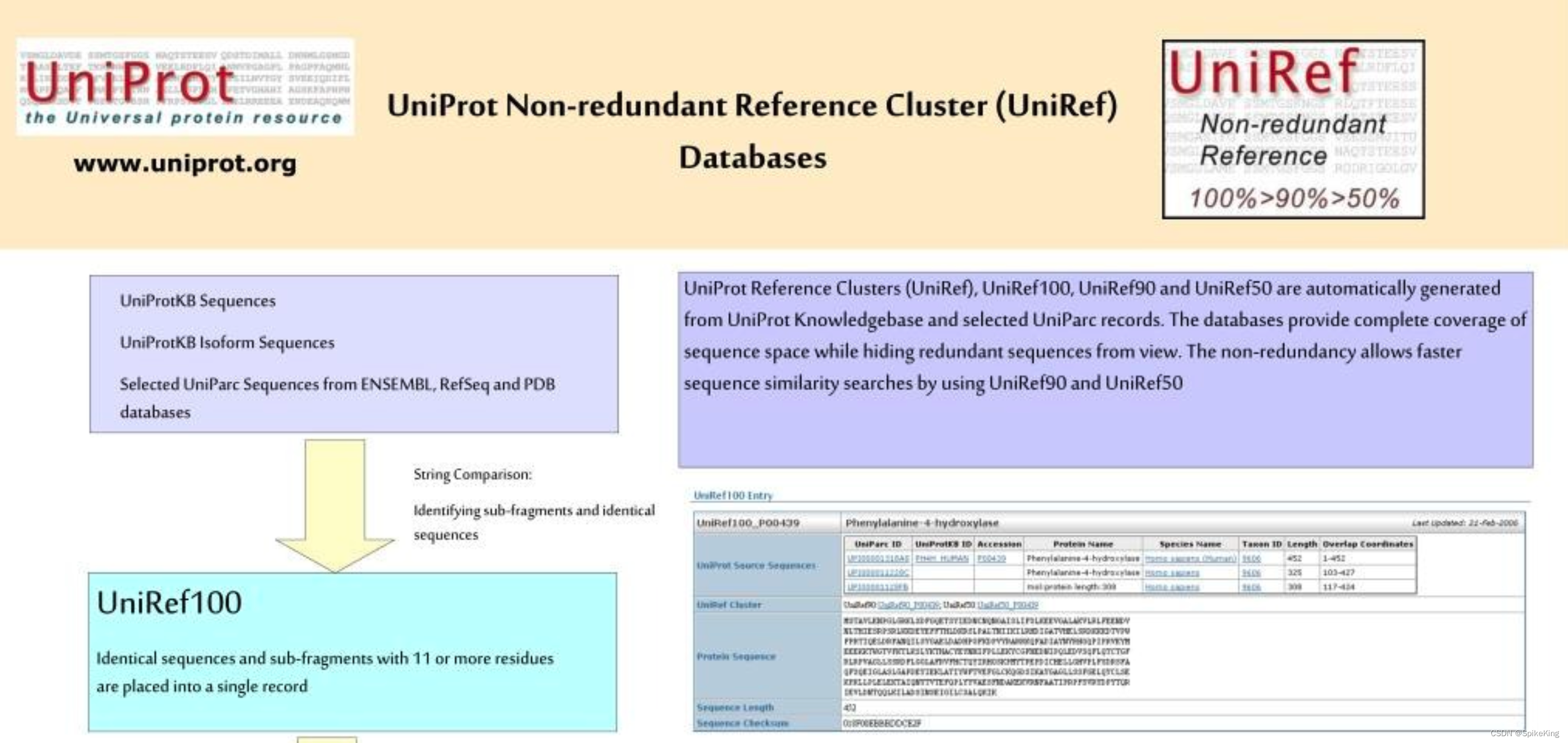
UniRef库:UniProt参考聚类(UniRef)的简称,提供了从UniProt知识库(包括异构体)和选定的UniParc记录中聚类得到的一组蛋白质序列,以便在不同的分辨率下覆盖整个序列空间,同时隐藏冗余的序列。UniRef库有4种分辨率:UniRef100、UniRef90、UniRef50、UniRef30,分别表示聚类的序列之间的相似度至少为100%、90%、50%、30%。UniRef库可以显著减少数据库的大小,提高序列相似性搜索的速度。UniRef库的代表性序列是根据以下优先级选择的:条目的质量、注释分数、物种、序列的长度。UniRef库是蛋白质序列计算领域的重要资源,可以用于蛋白质功能注释、进化分析、同源建模等应用。
UniRef序列中的物种(Species)信息:指每个聚类的代表性序列所属的物种,可以帮助用户了解聚类的生物学背景和多样性。物种信息是根据代表性序列的 UniProtKB 或 UniParc 条目中的物种注释获取的。如果一个聚类包含多个物种,那么物种信息会显示为“多物种聚类”。
其中,AlphaFold2 - Multimer 中,仅提供 uniprot_hits.sto 文件物种信息的提取,本文增加 uniref90_hits.sto 与 bfd_uniclust_hits.a3m 物种信息的提取,部分参考 DeepMSA2 的源码。
测试样本:8gmn_A276_B273_C264_D264_1077/msas/A,已搜索的 MSA 文件,如下:
642K bfd_uniclust_hits.a3m
26M mgnify_hits.sto
900M uniprot_hits.sto
701M uniref90_hits.sto
其中 mgnify_hits.sto 中,不包含物种信息,只有 UniRef 的搜索文件,才包括物种 (Species) 信息。
序列:
>chain_A
SIENGKYTCEEPYYYMEGVNVLGPKCVPVCGVPREPFEEKIIGGSDADIKNFPWQVFFDNPWAGGALINEYWVLTAAHVVEGNREPTMYVGSTSVQTSRLAKMLTPEHVFIHPGWKLLAVPEGRTNFDNDIALVRLKDPVKMGPTVSPICLPGTSSDYNLMDGDLGLISGWGRTEKRDRAVRLKAARLPVAPLRKCKEVKVEAYVFTPNMICAGGEKGMDSCKGDSGGAFAVQDPNDKTKFYAAGLVSWGPQCGTYGLYTRVKNYVDWIMKTMQEN
在 AF2 的默认配置,只使用 uniprot_hits.sto 计算物种信息,进行 MSA Pairing,可以扩展至全部的 MSA 文件:
| MSA | uniprot_hits | uniref90_hits | bfd_uniclust_hits |
|---|---|---|---|
| 格式 | sto | sto | msa |
| 文件 | 900M | 701M | 642K |
| 深度 | 124700 | 97958 | 1863 |
| 物种数量 | 1714/124700 | 4721/97958 | 164/351 |
| 物种示例 | CHACN | Scophthalmus maximus (物种ID 52904) | HAELO |
uniref90 物种信息,即,与 uniprot_hits 类似,物种 CHACN:
#=GS UniRef90_A0A6J2VG70/25-267 DE [subseq from] Uncharacterized protein LOC115812568 n=1 Tax=Chanos chanos TaxID=29144 RepID=A0A6J2VG70_CHACN
建议使用 TaxID 的 ID 作为物种的唯一标识。
其中 uniprot 与 uniclust 的物种信息,使用 正则表达式 提取:
# Sequences coming from UniProtKB database come in the
# `db|UniqueIdentifier|EntryName` format, e.g. `tr|A0A146SKV9|A0A146SKV9_FUNHE`
# or `sp|P0C2L1|A3X1_LOXLA` (for TREMBL/Swiss-Prot respectively).
UNIPROT_PATTERN = re.compile(r"""^# UniProtKB/TrEMBL or UniProtKB/Swiss-Prot(?:tr|sp)\|# A primary accession number of the UniProtKB entry.(?P<AccessionIdentifier>[A-Za-z0-9]{6,10})# Occasionally there is a _0 or _1 isoform suffix, which we ignore.(?:_\d)?\|# TREMBL repeats the accession ID here. Swiss-Prot has a mnemonic# protein ID code.(?:[A-Za-z0-9]+)_# A mnemonic species identification code.(?P<SpeciesIdentifier>([A-Za-z0-9]){1,5})# Small BFD uses a final value after an underscore, which we ignore.(?:_\d+)?$""",re.VERBOSE)# Sequences coming from Uniref database come in the
# `RepID=xxx_xxx` format, e.g. `RepID=A0A660T7U3_9SPIR`
# or `RepID=UPI00068451ED`
UNIREF_PATTERN = re.compile(r"""^# Uniref(RepID=)# A primary accession number of the UniProtKB entry.(?P<AccessionIdentifier>[A-Za-z0-9]{6,10})_# A mnemonic species identification code.(?P<SpeciesIdentifier>([A-Za-z0-9]){1,5})$""",re.VERBOSE)
源码:
#!/usr/bin/env python
# -- coding: utf-8 --
"""
Copyright (c) 2022. All rights reserved.
Created by C. L. Wang on 2024/1/15
"""import collections
import os
import re
import sysp = os.path.dirname(os.path.dirname(os.path.abspath(__file__)))
if p not in sys.path:sys.path.append(p)from root_dir import DATA_DIR# Sequences coming from UniProtKB database come in the
# `db|UniqueIdentifier|EntryName` format, e.g. `tr|A0A146SKV9|A0A146SKV9_FUNHE`
# or `sp|P0C2L1|A3X1_LOXLA` (for TREMBL/Swiss-Prot respectively).
UNIPROT_PATTERN = re.compile(r"""^# UniProtKB/TrEMBL or UniProtKB/Swiss-Prot(?:tr|sp)\|# A primary accession number of the UniProtKB entry.(?P<AccessionIdentifier>[A-Za-z0-9]{6,10})# Occasionally there is a _0 or _1 isoform suffix, which we ignore.(?:_\d)?\|# TREMBL repeats the accession ID here. Swiss-Prot has a mnemonic# protein ID code.(?:[A-Za-z0-9]+)_# A mnemonic species identification code.(?P<SpeciesIdentifier>([A-Za-z0-9]){1,5})# Small BFD uses a final value after an underscore, which we ignore.(?:_\d+)?$""",re.VERBOSE)# Sequences coming from Uniref database come in the
# `RepID=xxx_xxx` format, e.g. `RepID=A0A660T7U3_9SPIR`
# or `RepID=UPI00068451ED`
UNIREF_PATTERN = re.compile(r"""^# Uniref(RepID=)# A primary accession number of the UniProtKB entry.(?P<AccessionIdentifier>[A-Za-z0-9]{6,10})_# A mnemonic species identification code.(?P<SpeciesIdentifier>([A-Za-z0-9]){1,5})$""",re.VERBOSE)class SequencePairingUtils(object):"""MSA Pairing 与 提取蛋白质序列的物种信息"""def __init__(self):pass@staticmethoddef parse_sequence_specie(msa_sequence_identifier: str, dbtype: str = 'uniprot'):"""Gets species from a msa sequence identifier.The sequence identifier has the format specified by_UNIPROT_TREMBL_ENTRY_NAME_PATTERN or _UNIPROT_SWISSPROT_ENTRY_NAME_PATTERN.An example of a sequence identifier: `tr|A0A146SKV9|A0A146SKV9_FUNHE`Args:msa_sequence_identifier: a sequence identifier.dbtype: msa file type.Returns:An `Identifiers` instance with species_id. Thesecan be empty in the case where no identifier was found."""if dbtype == 'uniref':matches = re.search(UNIREF_PATTERN, msa_sequence_identifier.strip())else:matches = re.search(UNIPROT_PATTERN, msa_sequence_identifier.strip())species_id = ''if matches:species_id = matches.group('SpeciesIdentifier')return species_id@staticmethoddef extract_sequence_ids(id: str):"""Extracts sequence identifier from description. Returns None if no match."""split_id = id.split()if split_id:if split_id[0].lower().__contains__('uniref'):# return split_id[0].partition('/')[0],'uniprot' ### only return uniprot IDreturn split_id[-1].partition('/')[0], 'uniref'else:return split_id[0].partition('/')[0], 'uniprot'else:return None, None@classmethoddef get_species(cls, id: str, def_dbtype: str = ""):"""get species from id"""sequence_identifier, dbtype = cls.extract_sequence_ids(id)# print(sequence_identifier,dbtype)if def_dbtype:dbtype = def_dbtypeif sequence_identifier is None:return ''else:# print(parse_sequence_specie(sequence_identifier,dbtype=dbtype))return cls.parse_sequence_specie(sequence_identifier, dbtype=dbtype)@classmethoddef parse_a3m(cls, a3m_path):"""解析 a3m 文件,同时,生成物种信息,主要针对于 uniref 库的搜索结果:param a3m_path: a3m 文件:return query_seq: 查询序列, str:return seq_names: list, msa seq 的 描述信息:return name_to_species: dict, {描述信息: 物种}:return name_to_sequence: dict, {描述信息: 序列}"""seq_names = []name_to_species = {} # (seq_id, specie)name_to_sequence = {} # (seq_id, sequence)in_seqs = []with open(a3m_path, 'r') as f:a3m_txt = f.read()a3m_blocks = ('\n' + a3m_txt).split('\n>')[1:]for a3m_block in a3m_blocks:a3m_strs = a3m_block.split('\n')seq_id = a3m_strs[0]a3m_seq = ''for seq in a3m_strs[1:]:a3m_seq += seqseq_names.append(seq_id)name_to_sequence[seq_id] = a3m_seqin_seqs.append(a3m_seq)specie = cls.get_species(seq_id)if specie != "": # 确保包含物种信息name_to_species[seq_id] = speciequery_seq = in_seqs[0]# query_seq: 查询序列, str# seq_names: list, msa seq 的 描述信息# name_to_species: dict, {描述信息: 物种}# name_to_sequence: dict, {描述信息: 序列}return query_seq, seq_names, name_to_species, name_to_sequence@classmethoddef parse_sto_for_uniprot(cls, sto_path):"""解析 sto 文件,支持 uniprot,生成物种信息:param sto_path: msa 文件:return query_seq: 查询序列, str:return seq_names: list, msa seq 的 描述信息:return name_to_species: dict, {描述信息: 物种}:return name_to_sequence: dict, {描述信息: 序列}"""with open(sto_path) as f:sto_string = f.read()name_to_sequence = collections.OrderedDict()for line in sto_string.splitlines():line = line.strip()if not line or line.startswith(('#', '//')):continuename, sequence = line.split()if name not in name_to_sequence:name_to_sequence[name] = ''name_to_sequence[name] += sequencemsa = []keep_columns = []query_seq = ""for seq_index, sequence in enumerate(name_to_sequence.values()):if seq_index == 0:# Gather the columns with gaps from the queryquery = sequencekeep_columns = [i for i, res in enumerate(query) if res != '-']query_seq = ''.join([sequence[c] for c in keep_columns])# Remove the columns with gaps in the query from all sequences.aligned_sequence = ''.join([sequence[c] for c in keep_columns])msa.append(aligned_sequence)seq_names = list(name_to_sequence.keys())[1:]msa = msa[1:]name_to_species = dict()for desc in seq_names:s = cls.get_species(desc, def_dbtype="uniprot")if s != "": # 确保包含物种信息name_to_species[desc] = sname_to_sequence = dict()for desc, seq in zip(seq_names, msa):name_to_sequence[desc] = seq# query_seq: 查询序列, str# seq_names: list, msa seq 的 描述信息# name_to_species: dict, {描述信息: 物种}# name_to_sequence: dict, {描述信息: 序列}return query_seq, seq_names, name_to_species, name_to_sequence@classmethoddef parse_sto_for_uniref(cls, sto_path):"""解析 sto 文件,支持 uniref,生成物种信息:param sto_path: msa 文件:return query_seq: 查询序列, str:return seq_names: list, msa seq 的 描述信息:return name_to_species: dict, {描述信息: 物种}:return name_to_sequence: dict, {描述信息: 序列}"""with open(sto_path) as f:sto_string = f.read()name_to_sequence = collections.OrderedDict()for line in sto_string.splitlines():line = line.strip()if not line or line.startswith(('#', '//')):continuename, sequence = line.split()if name not in name_to_sequence:name_to_sequence[name] = ''name_to_sequence[name] += sequencemsa = []keep_columns = []query_seq = ""for seq_index, sequence in enumerate(name_to_sequence.values()):if seq_index == 0:# Gather the columns with gaps from the queryquery = sequencekeep_columns = [i for i, res in enumerate(query) if res != '-']query_seq = ''.join([sequence[c] for c in keep_columns])# Remove the columns with gaps in the query from all sequences.aligned_sequence = ''.join([sequence[c] for c in keep_columns])msa.append(aligned_sequence)# 解析物种name_to_species = collections.OrderedDict()for line in sto_string.splitlines():line = line.strip()if not line.startswith("#=GS"): # 只处理序列信息continueitems = line.split()name = items[1] # i.e. UniRef90_UPI001FA938EF/25-266# TaxID=52904for item in items:if item.startswith("TaxID"):species = item.split("=")[1]name_to_species[name] = speciesbreakseq_names = list(name_to_sequence.keys())[1:]msa = msa[1:]name_to_sequence = dict()for desc, seq in zip(seq_names, msa):name_to_sequence[desc] = seq# query_seq: 查询序列, str# seq_names: list, msa seq 的 描述信息# name_to_species: dict, {描述信息: 物种}# name_to_sequence: dict, {描述信息: 序列}return query_seq, seq_names, name_to_species, name_to_sequencedef process(self, input_path):print(f"[Info] input_path: {input_path}")assert os.path.isfile(input_path)if input_path.endswith("a3m"):query_seq, seq_names, name_to_species, name_to_sequence = self.parse_a3m(input_path)elif input_path.endswith("sto"):file_name = os.path.basename(input_path)if "uniprot" in file_name:query_seq, seq_names, name_to_species, name_to_sequence = self.parse_sto_for_uniprot(input_path)else:query_seq, seq_names, name_to_species, name_to_sequence = self.parse_sto_for_uniref(input_path)else:raise Exception(f"[Error] The input_path must be a3m or sto file ! {input_path}")print(f"[Info] species_set: {len(set(list(name_to_species.values())))}")print(f"[Info] query_seq: {query_seq}")print(f"[Info] seq_names: {len(seq_names)}, {seq_names[1]}")print(f"[Info] name_to_species: {len(name_to_species)}") # 字典 (key序列):(value物种)print(f"[Info] name_to_sequence: {len(name_to_sequence)}")for k in name_to_species.keys():s = name_to_species[k]if s:print(f"[Info] s: {s}, k: {k}")breakprint(f"[Info] 计算完成!")def main():spu = SequencePairingUtils()# name_to_species: 1863# species_set: 165, s: HAELO, tr|O96088|O96088_HAELO Serin proteinase 1# input_path = os.path.join(DATA_DIR, "templates", "msa", "bfd_uniclust_hits.a3m")# species_set: 1714, name_to_species: 124700# specie_set: 1715, s: CHACN, k: tr|A0A6J2VG70|A0A6J2VG70_CHACN/25-267# input_path = os.path.join(DATA_DIR, "templates", "msa", "uniprot_hits.sto")# species_set: 4721, seq_names: 97958input_path = os.path.join(DATA_DIR, "templates", "msa", "uniref90_hits.sto")spu.process(input_path)if __name__ == '__main__':main()
![[力扣 Hot100]Day7 接雨水](http://pic.xiahunao.cn/[力扣 Hot100]Day7 接雨水)


)











)

实现自动网络配置)

