大家好,欢迎来的单细胞图片美化专辑
1.如何修改seruat对象的行名 2.FeaturePlot如何把所有阳性表达的spot放到图的前面
在单细胞实践中,我发现不同的客户对画图需求并不一致,这可能和个人审美有关吧。本专辑着重于各种各样的单细胞个性化绘图。首先是pbmc数据下载,和标准处理流程,然后是本篇的个性化绘图。
读取pbmc数据
.libPaths(c( "/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2","/home/data/t040413/R/yll/usr/local/lib/R/site-library","/home/data/refdir/Rlib/", "/usr/local/lib/R/library"))getwd()dir.create('~/gzh/featureplot_dotplot_vlnplot')setwd('~/gzh/featureplot_dotplot_vlnplot')library(dplyr)library(Seurat)library(patchwork)pbmc=readRDS("~/gzh/featureplot_dotplot_vlnplot/pbmc3k_final.rds")DimPlot(pbmc,label = TRUE)
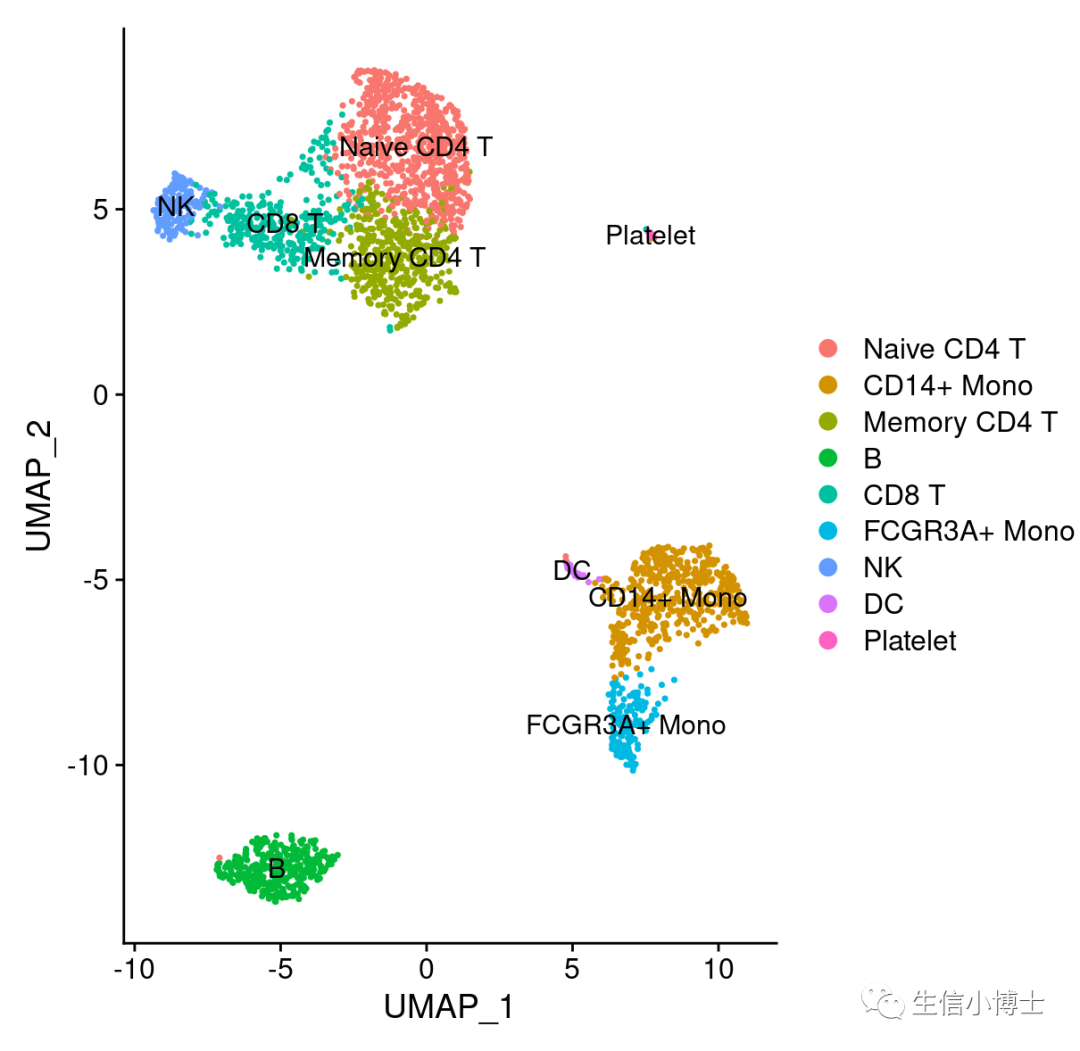
-
最基础的patchwork组合图
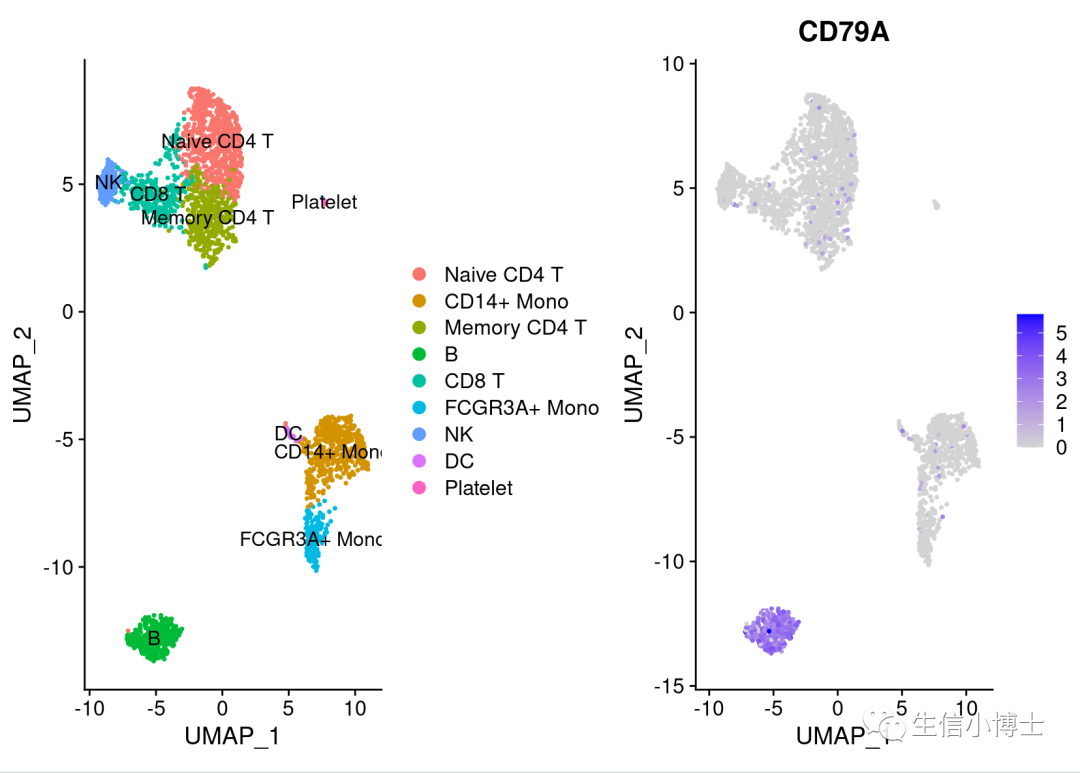
library(patchwork)DimPlot(pbmc,label = TRUE)|FeaturePlot(pbmc,features = 'CD79A')
2. featureplot个性化绘图 美化
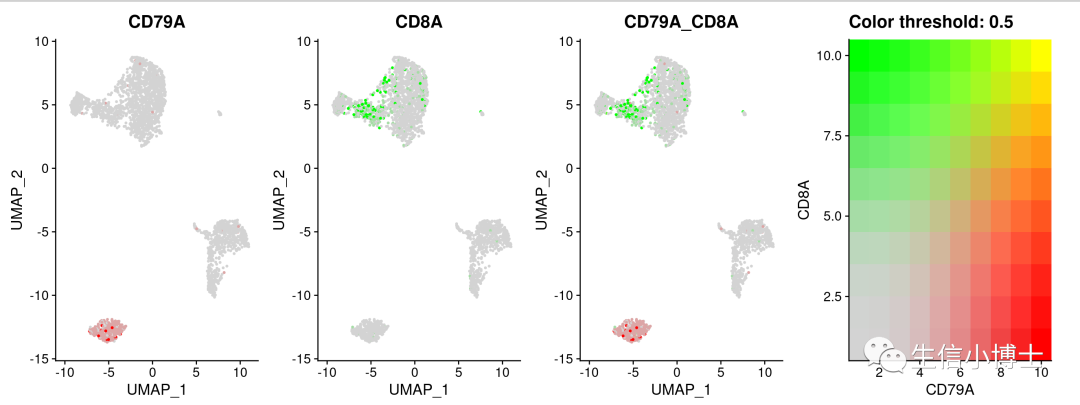
2 #featureplotFeaturePlot(pbmc,features = c('CD79A','CD8A'),blend=TRUE)
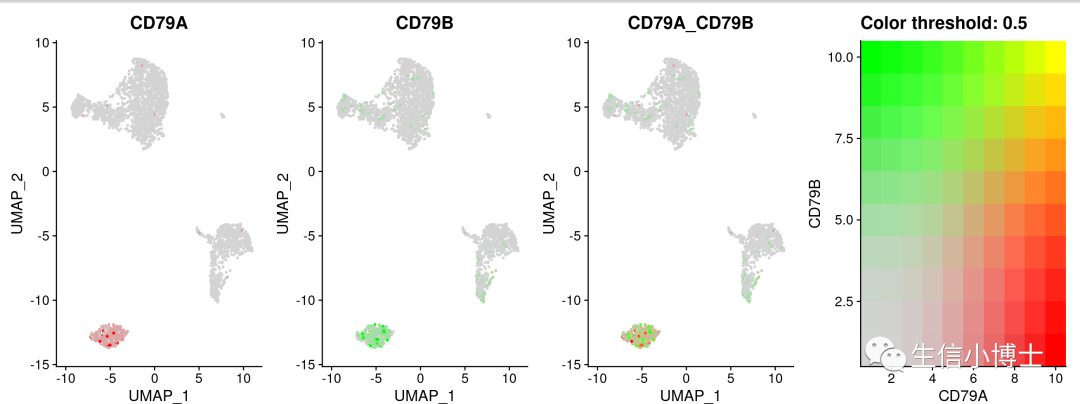
2 #featureplot
FeaturePlot(pbmc,features = c('CD79A','CD79B'),blend=TRUE)
2. 一次性画多张featureplot
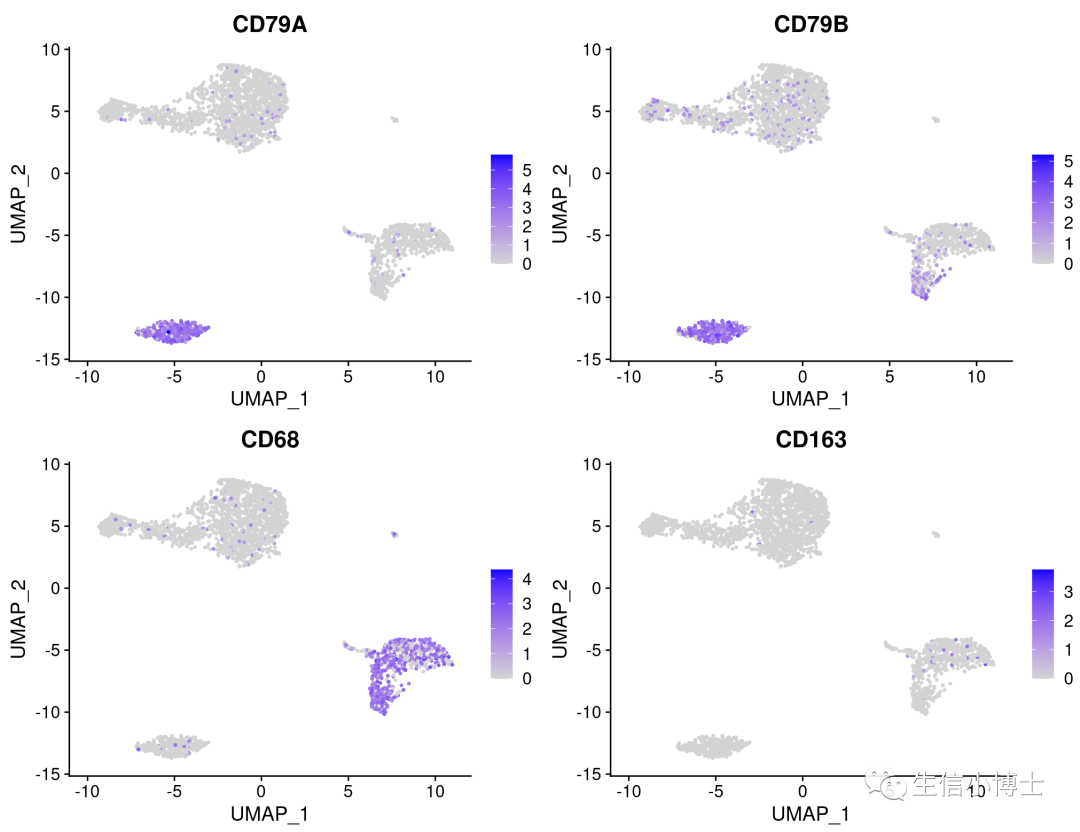
FeaturePlot(pbmc,features = c('CD79A','CD79B','CD68','CD163'))
2.1 featureplot个性化绘图 美化 等高线图 密度图,使用ggplot2来修改(二维密度的彩色等高线图)

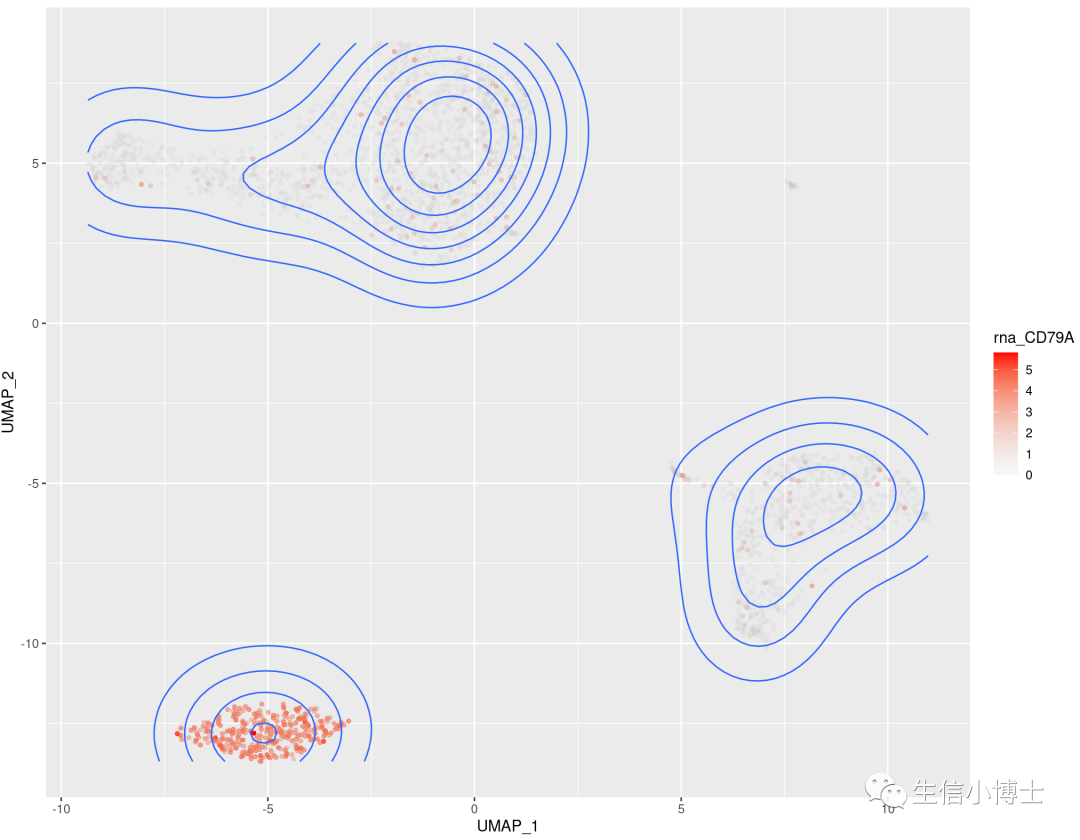
2 #featureplotFeaturePlot(pbmc,features = c('CD79A','CD79B'),blend=TRUE)2.1#library(ggplot2)library(ggthemes)library(ggnewscale)library(dplyr)head(pbmc@meta.data)mydata=FetchData(pbmc,vars = c('rna_CD79A','rna_CD8A','rna_CCR7','UMAP_1','UMAP_2'))head(mydata)# 绘制一个基因p <- ggplot(mydata,aes(x=UMAP_1,y=UMAP_2))+geom_point(data = mydata,aes(x=UMAP_1,y=UMAP_2,color=rna_CD79A),size=1)+scale_color_gradient('rna_CD79A',low = alpha('grey',0.1),#防止覆盖,把灰色的点透明度设置的高一些high = alpha('red',1))# 添加密度图p+stat_density2d(aes(colour=rna_CD79A))
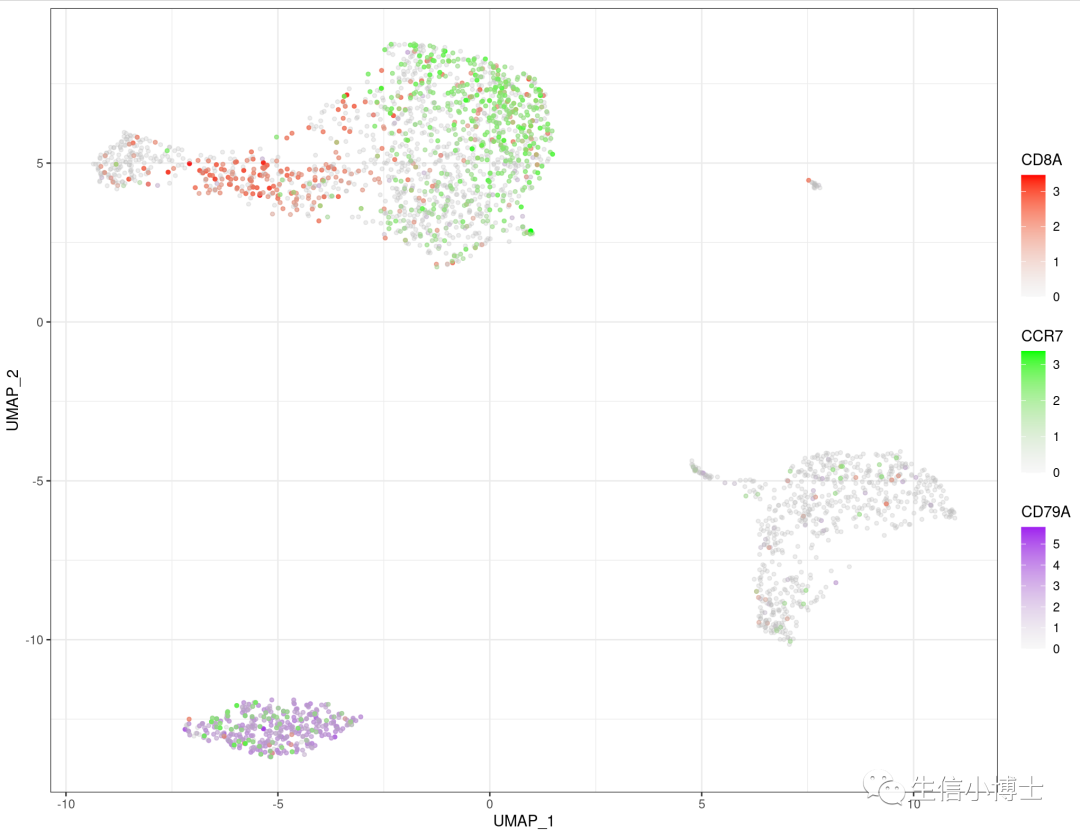
2.1 featureplot同时绘制多个基因
# 同时绘制三个基因
p <- ggplot(mydata,aes(x=UMAP_1,y=UMAP_2))+
geom_point(data = mydata,aes(x=UMAP_1,y=UMAP_2,
color=rna_CD79A),size=1)+
scale_color_gradient('CD79A',low = alpha('grey',0.1),#防止覆盖,把灰色的点透明度设置的高一些
high = alpha('purple',1))+#自己喜欢的颜色
new_scale('color')+
geom_point(data = mydata,aes(x=UMAP_1,y=UMAP_2,
color=rna_CD8A),size=1)+
scale_color_gradient('CD8A',low = alpha('grey',0.1),
high = alpha('red',1))+
new_scale('color')+
geom_point(data = mydata,aes(x=UMAP_1,y=UMAP_2,
color=rna_CCR7),size=1)+
scale_color_gradient( 'CCR7',low = alpha('grey',0.1),
high = alpha('green',1))+
theme_bw()
p
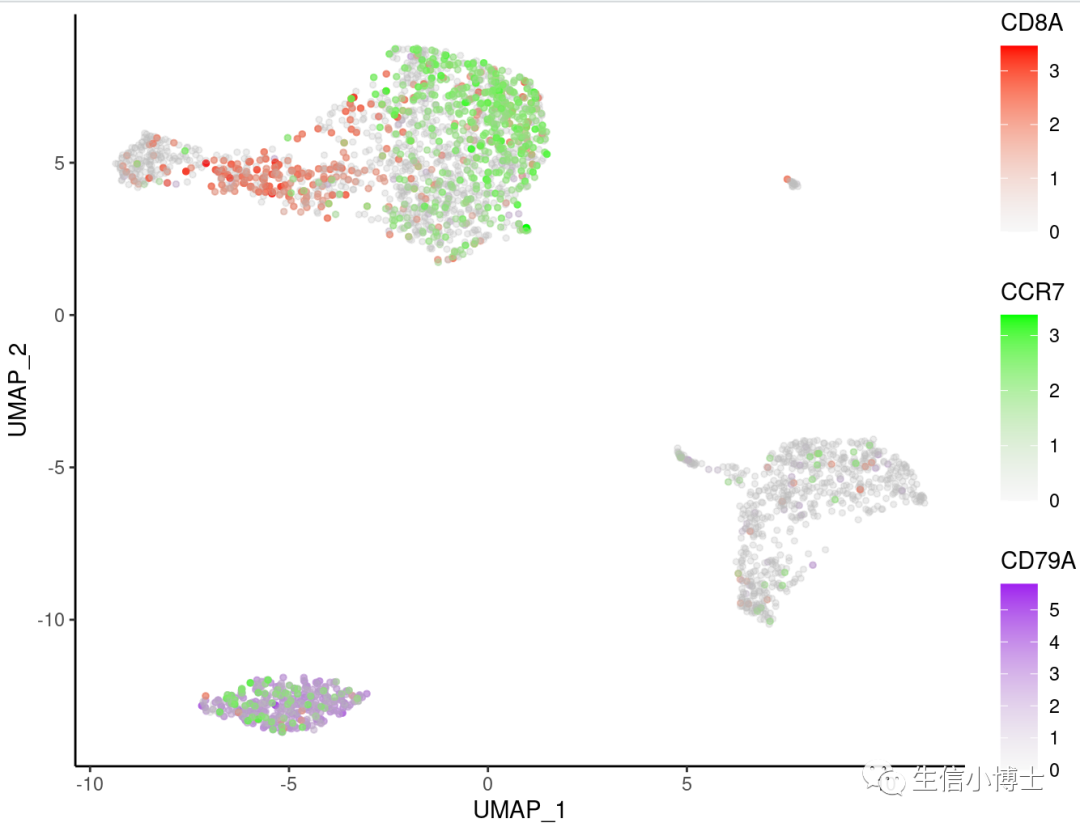
如果你想改ggplot背景的话
p+theme_bw()+theme_few()+
theme_classic()




问题)
)













