目录
简述
介绍 / 布置
加载数据
可视化数据集
建立 CNN
纠正数据失衡
训练模型
拟合模型
可视化模型性能
编辑预测和评估结果
政安晨的个人主页:政安晨
欢迎 👍点赞✍评论⭐收藏
收录专栏: TensorFlow与Keras机器学习实战
希望政安晨的博客能够对您有所裨益,如有不足之处,欢迎在评论区提出指正!
本文目标:基于 TPU 的医学图像分类。
简述
Keras是一个高级神经网络库,可以用于实现医学图像分类任务。医学图像分类是指将医学图像分为不同的类别,例如正常和异常,不同病种等。
在Keras中,可以使用卷积神经网络(CNN)来进行医学图像分类。CNN是一种特别适用于图像分类任务的神经网络架构,它能够有效地提取图像中的特征。
首先,需要准备医学图像数据集。可以使用公开的医学图像数据集,例如MNIST(手写数字图像)或者ImageNet(包含各种物体的图像)。另外,还可以使用自己收集的医学图像数据集。
接下来,需要定义CNN模型。可以使用Keras提供的各种层(例如卷积层、池化层、全连接层)来构建模型。卷积层可以有效地提取图像中的局部特征,池化层可以降低特征图的尺寸,全连接层可以用于最终的分类。
然后,需要编译模型。可以选择合适的损失函数和优化器来训练模型。对于多分类问题,可以使用交叉熵损失函数,常见的优化器包括Adam、SGD等。
接下来,需要进行模型训练。可以使用数据集中的图像进行训练,同时还需要准备对应的标签(即图像所属的类别)。可以使用Keras提供的fit函数来进行训练。
最后,可以使用模型进行预测。可以输入新的医学图像数据,使用模型预测其所属的类别。可以使用Keras提供的predict函数来进行预测。
总的来说,Keras提供了一个简单且强大的工具,可以用于医学图像分类任务。通过定义、编译、训练和预测模型,可以提高医学图像分类的准确性和效率,为医学诊断和研究提供有力支持。
介绍 / 布置
本文将介绍如何建立 X 光图像分类模型,以预测 X 光扫描是否显示存在肺炎。
import re
import os
import random
import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plttry:tpu = tf.distribute.cluster_resolver.TPUClusterResolver.connect()print("Device:", tpu.master())strategy = tf.distribute.TPUStrategy(tpu)
except:strategy = tf.distribute.get_strategy()
print("Number of replicas:", strategy.num_replicas_in_sync)演绎:
Device: grpc://10.0.27.122:8470
INFO:tensorflow:Initializing the TPU system: grpc://10.0.27.122:8470INFO:tensorflow:Initializing the TPU system: grpc://10.0.27.122:8470INFO:tensorflow:Clearing out eager cachesINFO:tensorflow:Clearing out eager cachesINFO:tensorflow:Finished initializing TPU system.INFO:tensorflow:Finished initializing TPU system.
WARNING:absl:[`tf.distribute.TPUStrategy`](https://www.tensorflow.org/api_docs/python/tf/distribute/TPUStrategy) is deprecated, please use the non experimental symbol [`tf.distribute.TPUStrategy`](https://www.tensorflow.org/api_docs/python/tf/distribute/TPUStrategy) instead.INFO:tensorflow:Found TPU system:INFO:tensorflow:Found TPU system:INFO:tensorflow:*** Num TPU Cores: 8INFO:tensorflow:*** Num TPU Cores: 8INFO:tensorflow:*** Num TPU Workers: 1INFO:tensorflow:*** Num TPU Workers: 1INFO:tensorflow:*** Num TPU Cores Per Worker: 8INFO:tensorflow:*** Num TPU Cores Per Worker: 8INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:localhost/replica:0/task:0/device:CPU:0, CPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:localhost/replica:0/task:0/device:CPU:0, CPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:localhost/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:localhost/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:CPU:0, CPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:CPU:0, CPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:0, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:0, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:1, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:1, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:2, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:2, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:3, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:3, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:4, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:4, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:5, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:5, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:6, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:6, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:7, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU:7, TPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU_SYSTEM:0, TPU_SYSTEM, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:TPU_SYSTEM:0, TPU_SYSTEM, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 0, 0)INFO:tensorflow:*** Available Device: _DeviceAttributes(/job:worker/replica:0/task:0/device:XLA_CPU:0, XLA_CPU, 0, 0)Number of replicas: 8我们需要谷歌云链接到我们的数据,以便使用 TPU 加载数据。下面,我们将定义本示例中使用的关键配置参数。要在 TPU 上运行,本示例必须在 Colab 上选择 TPU 运行时。
AUTOTUNE = tf.data.AUTOTUNE
BATCH_SIZE = 25 * strategy.num_replicas_in_sync
IMAGE_SIZE = [180, 180]
CLASS_NAMES = ["NORMAL", "PNEUMONIA"]加载数据
我们使用的 Cell 胸部 X 光数据将数据分为训练文件和测试文件。首先加载训练 TFRecords。
train_images = tf.data.TFRecordDataset("gs://download.tensorflow.org/data/ChestXRay2017/train/images.tfrec"
)
train_paths = tf.data.TFRecordDataset("gs://download.tensorflow.org/data/ChestXRay2017/train/paths.tfrec"
)ds = tf.data.Dataset.zip((train_images, train_paths))让我们数一数有多少张健康/正常的胸部 X 光片,有多少张肺炎胸部 X 光片:
COUNT_NORMAL = len([filenamefor filename in train_pathsif "NORMAL" in filename.numpy().decode("utf-8")]
)
print("Normal images count in training set: " + str(COUNT_NORMAL))COUNT_PNEUMONIA = len([filenamefor filename in train_pathsif "PNEUMONIA" in filename.numpy().decode("utf-8")]
)
print("Pneumonia images count in training set: " + str(COUNT_PNEUMONIA))结果:
Normal images count in training set: 1349
Pneumonia images count in training set: 3883请注意,被归类为肺炎的图像要比正常图像多得多。这说明我们的数据不平衡。我们稍后将在笔记本中纠正这种不平衡。
我们要将每个文件名映射到相应的(图像、标签)对。以下方法将帮助我们实现这一目标。
由于只有两个标签,我们将对标签进行编码,使 1 或 True 表示肺炎,0 或 False 表示正常。
def get_label(file_path):# convert the path to a list of path componentsparts = tf.strings.split(file_path, "/")# The second to last is the class-directoryif parts[-2] == "PNEUMONIA":return 1else:return 0def decode_img(img):# convert the compressed string to a 3D uint8 tensorimg = tf.image.decode_jpeg(img, channels=3)# resize the image to the desired size.return tf.image.resize(img, IMAGE_SIZE)def process_path(image, path):label = get_label(path)# load the raw data from the file as a stringimg = decode_img(image)return img, labelds = ds.map(process_path, num_parallel_calls=AUTOTUNE)让我们把数据分成训练数据集和验证数据集。
ds = ds.shuffle(10000)
train_ds = ds.take(4200)
val_ds = ds.skip(4200)让我们来想象一下(图像、标签)对的形状。
for image, label in train_ds.take(1):print("Image shape: ", image.numpy().shape)print("Label: ", label.numpy())结果:
Image shape: (180, 180, 3)
Label: False同时加载并格式化测试数据。
test_images = tf.data.TFRecordDataset("gs://download.tensorflow.org/data/ChestXRay2017/test/images.tfrec"
)
test_paths = tf.data.TFRecordDataset("gs://download.tensorflow.org/data/ChestXRay2017/test/paths.tfrec"
)
test_ds = tf.data.Dataset.zip((test_images, test_paths))test_ds = test_ds.map(process_path, num_parallel_calls=AUTOTUNE)
test_ds = test_ds.batch(BATCH_SIZE)可视化数据集
首先,让我们使用缓冲预取,这样就能从磁盘获取数据,而不会出现 I/O 阻塞。
请注意,大型图像数据集不应缓存在内存中。我们在这里这样做是因为数据集不是很大,而且我们想在 TPU 上进行训练。
def prepare_for_training(ds, cache=True):# This is a small dataset, only load it once, and keep it in memory.# use `.cache(filename)` to cache preprocessing work for datasets that don't# fit in memory.if cache:if isinstance(cache, str):ds = ds.cache(cache)else:ds = ds.cache()ds = ds.batch(BATCH_SIZE)# `prefetch` lets the dataset fetch batches in the background while the model# is training.ds = ds.prefetch(buffer_size=AUTOTUNE)return ds调用下一批迭代训练数据。
train_ds = prepare_for_training(train_ds)
val_ds = prepare_for_training(val_ds)image_batch, label_batch = next(iter(train_ds))定义在批次中显示图像的方法。
def show_batch(image_batch, label_batch):plt.figure(figsize=(10, 10))for n in range(25):ax = plt.subplot(5, 5, n + 1)plt.imshow(image_batch[n] / 255)if label_batch[n]:plt.title("PNEUMONIA")else:plt.title("NORMAL")plt.axis("off")由于该方法将 NumPy 数组作为参数,因此在批次上调用 numpy 函数,以 NumPy 数组形式返回张量。
show_batch(image_batch.numpy(), label_batch.numpy())
建立 CNN
为了使我们的模型更加模块化和易于理解,让我们定义一些模块。在构建卷积神经网络时,我们将创建一个卷积块和一个密集层块。
import os
os.environ['KERAS_BACKEND'] = 'tensorflow'import keras
from keras import layersdef conv_block(filters, inputs):x = layers.SeparableConv2D(filters, 3, activation="relu", padding="same")(inputs)x = layers.SeparableConv2D(filters, 3, activation="relu", padding="same")(x)x = layers.BatchNormalization()(x)outputs = layers.MaxPool2D()(x)return outputsdef dense_block(units, dropout_rate, inputs):x = layers.Dense(units, activation="relu")(inputs)x = layers.BatchNormalization()(x)outputs = layers.Dropout(dropout_rate)(x)return outputs下面的方法将定义为我们建立模型的函数。
图像的原始值范围为 [0,255]。CNN 在使用较小的数值时效果更好,因此我们将缩小输入范围。
剔除层非常重要,因为它们可以降低模型过拟合的可能性。我们希望以一个节点的密集层结束模型,因为这将是决定 X 光片是否显示肺炎的二进制输出。
def build_model():inputs = keras.Input(shape=(IMAGE_SIZE[0], IMAGE_SIZE[1], 3))x = layers.Rescaling(1.0 / 255)(inputs)x = layers.Conv2D(16, 3, activation="relu", padding="same")(x)x = layers.Conv2D(16, 3, activation="relu", padding="same")(x)x = layers.MaxPool2D()(x)x = conv_block(32, x)x = conv_block(64, x)x = conv_block(128, x)x = layers.Dropout(0.2)(x)x = conv_block(256, x)x = layers.Dropout(0.2)(x)x = layers.Flatten()(x)x = dense_block(512, 0.7, x)x = dense_block(128, 0.5, x)x = dense_block(64, 0.3, x)outputs = layers.Dense(1, activation="sigmoid")(x)model = keras.Model(inputs=inputs, outputs=outputs)return model纠正数据失衡
在本示例的前面部分,我们看到数据是不平衡的,被归类为肺炎的图像多于正常图像。
我们将通过类别加权来纠正这种情况:
initial_bias = np.log([COUNT_PNEUMONIA / COUNT_NORMAL])
print("Initial bias: {:.5f}".format(initial_bias[0]))TRAIN_IMG_COUNT = COUNT_NORMAL + COUNT_PNEUMONIA
weight_for_0 = (1 / COUNT_NORMAL) * (TRAIN_IMG_COUNT) / 2.0
weight_for_1 = (1 / COUNT_PNEUMONIA) * (TRAIN_IMG_COUNT) / 2.0class_weight = {0: weight_for_0, 1: weight_for_1}print("Weight for class 0: {:.2f}".format(weight_for_0))
print("Weight for class 1: {:.2f}".format(weight_for_1))结果如下:
Initial bias: 1.05724
Weight for class 0: 1.94
Weight for class 1: 0.67类别 0(正常)的权重远远高于类别 1(肺炎)的权重。因为正常图像的数量较少,所以每个正常图像的权重会更高,以平衡数据,因为 CNN 在训练数据平衡的情况下工作效果最佳。
训练模型
定义回调
检查点回调会保存模型的最佳权重,这样下次我们想使用模型时,就不必再花时间训练它了。早期停止回调会在模型开始停滞不前,或者更糟糕的是模型开始过度拟合时停止训练过程。
checkpoint_cb = keras.callbacks.ModelCheckpoint("xray_model.keras", save_best_only=True)early_stopping_cb = keras.callbacks.EarlyStopping(patience=10, restore_best_weights=True
)我们还需要调整学习率。学习率太高会导致模型发散。学习率太小会导致模型太慢。我们将在下文中采用指数学习率调度方法。
initial_learning_rate = 0.015
lr_schedule = keras.optimizers.schedules.ExponentialDecay(initial_learning_rate, decay_steps=100000, decay_rate=0.96, staircase=True
)拟合模型
对于我们的指标,我们希望包括精确度和召回率,因为它们能让我们更清楚地了解我们的模型有多好。精确度告诉我们有多少标签是正确的。由于我们的数据并不均衡,因此准确率可能会对一个好的模型产生偏差(例如,一个总是预测 PNEUMONIA 的模型准确率为 74%,但并不是一个好的模型)。
精确度是真阳性(TP)与假阳性(FP)之和的比值。它显示了标签阳性结果中真正正确的比例。
Recall 是 TP 与假阴性(FN)之和的 TP 数。它显示了实际阳性结果中正确率的百分比。
由于图像只有两种可能的标签,我们将使用二元交叉熵损失。在拟合模型时,请记住指定我们之前定义的类权重。因为我们使用的是 TPU,所以训练时间会很快,不到 2 分钟。
with strategy.scope():model = build_model()METRICS = [keras.metrics.BinaryAccuracy(),keras.metrics.Precision(name="precision"),keras.metrics.Recall(name="recall"),]model.compile(optimizer=keras.optimizers.Adam(learning_rate=lr_schedule),loss="binary_crossentropy",metrics=METRICS,)history = model.fit(train_ds,epochs=100,validation_data=val_ds,class_weight=class_weight,callbacks=[checkpoint_cb, early_stopping_cb],
)Epoch 1/100
WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/data/ops/multi_device_iterator_ops.py:601: get_next_as_optional (from tensorflow.python.data.ops.iterator_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.data.Iterator.get_next_as_optional()` instead.WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/data/ops/multi_device_iterator_ops.py:601: get_next_as_optional (from tensorflow.python.data.ops.iterator_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use `tf.data.Iterator.get_next_as_optional()` instead.21/21 [==============================] - 12s 568ms/step - loss: 0.5857 - binary_accuracy: 0.6960 - precision: 0.8887 - recall: 0.6733 - val_loss: 34.0149 - val_binary_accuracy: 0.7180 - val_precision: 0.7180 - val_recall: 1.0000
Epoch 2/100
21/21 [==============================] - 3s 128ms/step - loss: 0.2916 - binary_accuracy: 0.8755 - precision: 0.9540 - recall: 0.8738 - val_loss: 97.5194 - val_binary_accuracy: 0.7180 - val_precision: 0.7180 - val_recall: 1.0000
Epoch 3/100
21/21 [==============================] - 4s 167ms/step - loss: 0.2384 - binary_accuracy: 0.9002 - precision: 0.9663 - recall: 0.8964 - val_loss: 27.7902 - val_binary_accuracy: 0.7180 - val_precision: 0.7180 - val_recall: 1.0000
Epoch 4/100
21/21 [==============================] - 4s 173ms/step - loss: 0.2046 - binary_accuracy: 0.9145 - precision: 0.9725 - recall: 0.9102 - val_loss: 10.8302 - val_binary_accuracy: 0.7180 - val_precision: 0.7180 - val_recall: 1.0000
Epoch 5/100
21/21 [==============================] - 4s 174ms/step - loss: 0.1841 - binary_accuracy: 0.9279 - precision: 0.9733 - recall: 0.9279 - val_loss: 3.5860 - val_binary_accuracy: 0.7103 - val_precision: 0.7162 - val_recall: 0.9879
Epoch 6/100
21/21 [==============================] - 4s 185ms/step - loss: 0.1600 - binary_accuracy: 0.9362 - precision: 0.9791 - recall: 0.9337 - val_loss: 0.3014 - val_binary_accuracy: 0.8895 - val_precision: 0.8973 - val_recall: 0.9555
Epoch 7/100
21/21 [==============================] - 3s 130ms/step - loss: 0.1567 - binary_accuracy: 0.9393 - precision: 0.9798 - recall: 0.9372 - val_loss: 0.6763 - val_binary_accuracy: 0.7810 - val_precision: 0.7760 - val_recall: 0.9771
Epoch 8/100
21/21 [==============================] - 3s 131ms/step - loss: 0.1532 - binary_accuracy: 0.9421 - precision: 0.9825 - recall: 0.9385 - val_loss: 0.3169 - val_binary_accuracy: 0.8895 - val_precision: 0.8684 - val_recall: 0.9973
Epoch 9/100
21/21 [==============================] - 4s 184ms/step - loss: 0.1457 - binary_accuracy: 0.9431 - precision: 0.9822 - recall: 0.9401 - val_loss: 0.2064 - val_binary_accuracy: 0.9273 - val_precision: 0.9840 - val_recall: 0.9136
Epoch 10/100
21/21 [==============================] - 3s 132ms/step - loss: 0.1201 - binary_accuracy: 0.9521 - precision: 0.9869 - recall: 0.9479 - val_loss: 0.4364 - val_binary_accuracy: 0.8605 - val_precision: 0.8443 - val_recall: 0.9879
Epoch 11/100
21/21 [==============================] - 3s 127ms/step - loss: 0.1200 - binary_accuracy: 0.9510 - precision: 0.9863 - recall: 0.9469 - val_loss: 0.5197 - val_binary_accuracy: 0.8508 - val_precision: 1.0000 - val_recall: 0.7922
Epoch 12/100
21/21 [==============================] - 4s 186ms/step - loss: 0.1077 - binary_accuracy: 0.9581 - precision: 0.9870 - recall: 0.9559 - val_loss: 0.1349 - val_binary_accuracy: 0.9486 - val_precision: 0.9587 - val_recall: 0.9703
Epoch 13/100
21/21 [==============================] - 4s 173ms/step - loss: 0.0918 - binary_accuracy: 0.9650 - precision: 0.9914 - recall: 0.9611 - val_loss: 0.0926 - val_binary_accuracy: 0.9700 - val_precision: 0.9837 - val_recall: 0.9744
Epoch 14/100
21/21 [==============================] - 3s 130ms/step - loss: 0.0996 - binary_accuracy: 0.9612 - precision: 0.9913 - recall: 0.9559 - val_loss: 0.1811 - val_binary_accuracy: 0.9419 - val_precision: 0.9956 - val_recall: 0.9231
Epoch 15/100
21/21 [==============================] - 3s 129ms/step - loss: 0.0898 - binary_accuracy: 0.9643 - precision: 0.9901 - recall: 0.9614 - val_loss: 0.1525 - val_binary_accuracy: 0.9486 - val_precision: 0.9986 - val_recall: 0.9298
Epoch 16/100
21/21 [==============================] - 3s 128ms/step - loss: 0.0941 - binary_accuracy: 0.9621 - precision: 0.9904 - recall: 0.9582 - val_loss: 0.5101 - val_binary_accuracy: 0.8527 - val_precision: 1.0000 - val_recall: 0.7949
Epoch 17/100
21/21 [==============================] - 3s 125ms/step - loss: 0.0798 - binary_accuracy: 0.9636 - precision: 0.9897 - recall: 0.9607 - val_loss: 0.1239 - val_binary_accuracy: 0.9622 - val_precision: 0.9875 - val_recall: 0.9595
Epoch 18/100
21/21 [==============================] - 3s 126ms/step - loss: 0.0821 - binary_accuracy: 0.9657 - precision: 0.9911 - recall: 0.9623 - val_loss: 0.1597 - val_binary_accuracy: 0.9322 - val_precision: 0.9956 - val_recall: 0.9096
Epoch 19/100
21/21 [==============================] - 3s 143ms/step - loss: 0.0800 - binary_accuracy: 0.9657 - precision: 0.9917 - recall: 0.9617 - val_loss: 0.2538 - val_binary_accuracy: 0.9109 - val_precision: 1.0000 - val_recall: 0.8758
Epoch 20/100
21/21 [==============================] - 3s 127ms/step - loss: 0.0605 - binary_accuracy: 0.9738 - precision: 0.9950 - recall: 0.9694 - val_loss: 0.6594 - val_binary_accuracy: 0.8566 - val_precision: 1.0000 - val_recall: 0.8003
Epoch 21/100
21/21 [==============================] - 4s 167ms/step - loss: 0.0726 - binary_accuracy: 0.9733 - precision: 0.9937 - recall: 0.9701 - val_loss: 0.0593 - val_binary_accuracy: 0.9816 - val_precision: 0.9945 - val_recall: 0.9798
Epoch 22/100
21/21 [==============================] - 3s 126ms/step - loss: 0.0577 - binary_accuracy: 0.9783 - precision: 0.9951 - recall: 0.9755 - val_loss: 0.1087 - val_binary_accuracy: 0.9729 - val_precision: 0.9931 - val_recall: 0.9690
Epoch 23/100
21/21 [==============================] - 3s 125ms/step - loss: 0.0652 - binary_accuracy: 0.9729 - precision: 0.9924 - recall: 0.9707 - val_loss: 1.8465 - val_binary_accuracy: 0.7180 - val_precision: 0.7180 - val_recall: 1.0000
Epoch 24/100
21/21 [==============================] - 3s 124ms/step - loss: 0.0538 - binary_accuracy: 0.9783 - precision: 0.9951 - recall: 0.9755 - val_loss: 1.5769 - val_binary_accuracy: 0.7180 - val_precision: 0.7180 - val_recall: 1.0000
Epoch 25/100
21/21 [==============================] - 4s 167ms/step - loss: 0.0549 - binary_accuracy: 0.9776 - precision: 0.9954 - recall: 0.9743 - val_loss: 0.0590 - val_binary_accuracy: 0.9777 - val_precision: 0.9904 - val_recall: 0.9784
Epoch 26/100
21/21 [==============================] - 3s 131ms/step - loss: 0.0677 - binary_accuracy: 0.9719 - precision: 0.9924 - recall: 0.9694 - val_loss: 2.6008 - val_binary_accuracy: 0.6928 - val_precision: 0.9977 - val_recall: 0.5735
Epoch 27/100
21/21 [==============================] - 3s 127ms/step - loss: 0.0469 - binary_accuracy: 0.9833 - precision: 0.9971 - recall: 0.9804 - val_loss: 1.0184 - val_binary_accuracy: 0.8605 - val_precision: 0.9983 - val_recall: 0.8070
Epoch 28/100
21/21 [==============================] - 3s 126ms/step - loss: 0.0501 - binary_accuracy: 0.9790 - precision: 0.9961 - recall: 0.9755 - val_loss: 0.3737 - val_binary_accuracy: 0.9089 - val_precision: 0.9954 - val_recall: 0.8772
Epoch 29/100
21/21 [==============================] - 3s 128ms/step - loss: 0.0548 - binary_accuracy: 0.9798 - precision: 0.9941 - recall: 0.9784 - val_loss: 1.2928 - val_binary_accuracy: 0.7907 - val_precision: 1.0000 - val_recall: 0.7085
Epoch 30/100
21/21 [==============================] - 3s 129ms/step - loss: 0.0370 - binary_accuracy: 0.9860 - precision: 0.9980 - recall: 0.9829 - val_loss: 0.1370 - val_binary_accuracy: 0.9612 - val_precision: 0.9972 - val_recall: 0.9487
Epoch 31/100
21/21 [==============================] - 3s 125ms/step - loss: 0.0585 - binary_accuracy: 0.9819 - precision: 0.9951 - recall: 0.9804 - val_loss: 1.1955 - val_binary_accuracy: 0.6870 - val_precision: 0.9976 - val_recall: 0.5655
Epoch 32/100
21/21 [==============================] - 3s 140ms/step - loss: 0.0813 - binary_accuracy: 0.9695 - precision: 0.9934 - recall: 0.9652 - val_loss: 1.0394 - val_binary_accuracy: 0.8576 - val_precision: 0.9853 - val_recall: 0.8138
Epoch 33/100
21/21 [==============================] - 3s 128ms/step - loss: 0.1111 - binary_accuracy: 0.9555 - precision: 0.9870 - recall: 0.9524 - val_loss: 4.9438 - val_binary_accuracy: 0.5911 - val_precision: 1.0000 - val_recall: 0.4305
Epoch 34/100
21/21 [==============================] - 3s 130ms/step - loss: 0.0680 - binary_accuracy: 0.9726 - precision: 0.9921 - recall: 0.9707 - val_loss: 2.8822 - val_binary_accuracy: 0.7267 - val_precision: 0.9978 - val_recall: 0.6208
Epoch 35/100
21/21 [==============================] - 4s 187ms/step - loss: 0.0784 - binary_accuracy: 0.9712 - precision: 0.9892 - recall: 0.9717 - val_loss: 0.3940 - val_binary_accuracy: 0.9390 - val_precision: 0.9942 - val_recall: 0.9204可视化模型性能
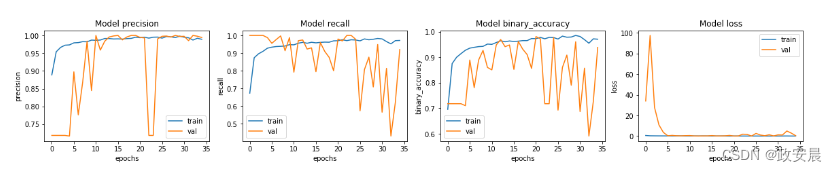
让我们绘制训练集和验证集的模型准确率和损失图。请注意,本笔记本没有指定随机种子。对于您的笔记本电脑,可能会有轻微差异。
fig, ax = plt.subplots(1, 4, figsize=(20, 3))
ax = ax.ravel()for i, met in enumerate(["precision", "recall", "binary_accuracy", "loss"]):ax[i].plot(history.history[met])ax[i].plot(history.history["val_" + met])ax[i].set_title("Model {}".format(met))ax[i].set_xlabel("epochs")ax[i].set_ylabel(met)ax[i].legend(["train", "val"])(我们看到,我们模型的准确率约为 95%。)
 预测和评估结果
预测和评估结果
让我们在测试数据上对模型进行评估!
model.evaluate(test_ds, return_dict=True)4/4 [==============================] - 3s 708ms/step - loss: 0.9718 - binary_accuracy: 0.7901 - precision: 0.7524 - recall: 0.9897{'binary_accuracy': 0.7900640964508057,'loss': 0.9717951416969299,'precision': 0.752436637878418,'recall': 0.9897436499595642}我们发现,测试数据的准确率低于验证集的准确率。这可能表明拟合过度。
我们的召回率大于精确率,这表明几乎所有肺炎图像都被正确识别,但一些正常图像被错误识别。我们应该努力提高精确度。
for image, label in test_ds.take(1):plt.imshow(image[0] / 255.0)plt.title(CLASS_NAMES[label[0].numpy()])prediction = model.predict(test_ds.take(1))[0]
scores = [1 - prediction, prediction]for score, name in zip(scores, CLASS_NAMES):print("This image is %.2f percent %s" % ((100 * score), name))执行如下:
/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:3: DeprecationWarning: In future, it will be an error for 'np.bool_' scalars to be interpreted as an indexThis is separate from the ipykernel package so we can avoid doing imports untilThis image is 47.19 percent NORMAL
This image is 52.81 percent PNEUMONIA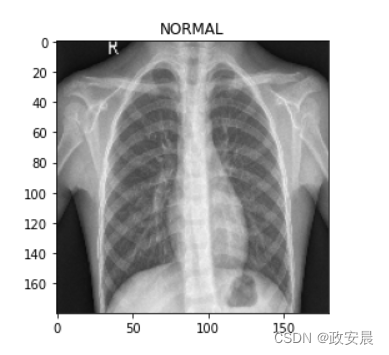
(FHQ-Treap版))




》第8章-系统质量属性与架构评估-03-ATAM方法架构评估实践(下))
)



)
:深入理解一维数组和二维数组)

 Object Pascal 学习笔记---第9章第1节(Try-Except块))





