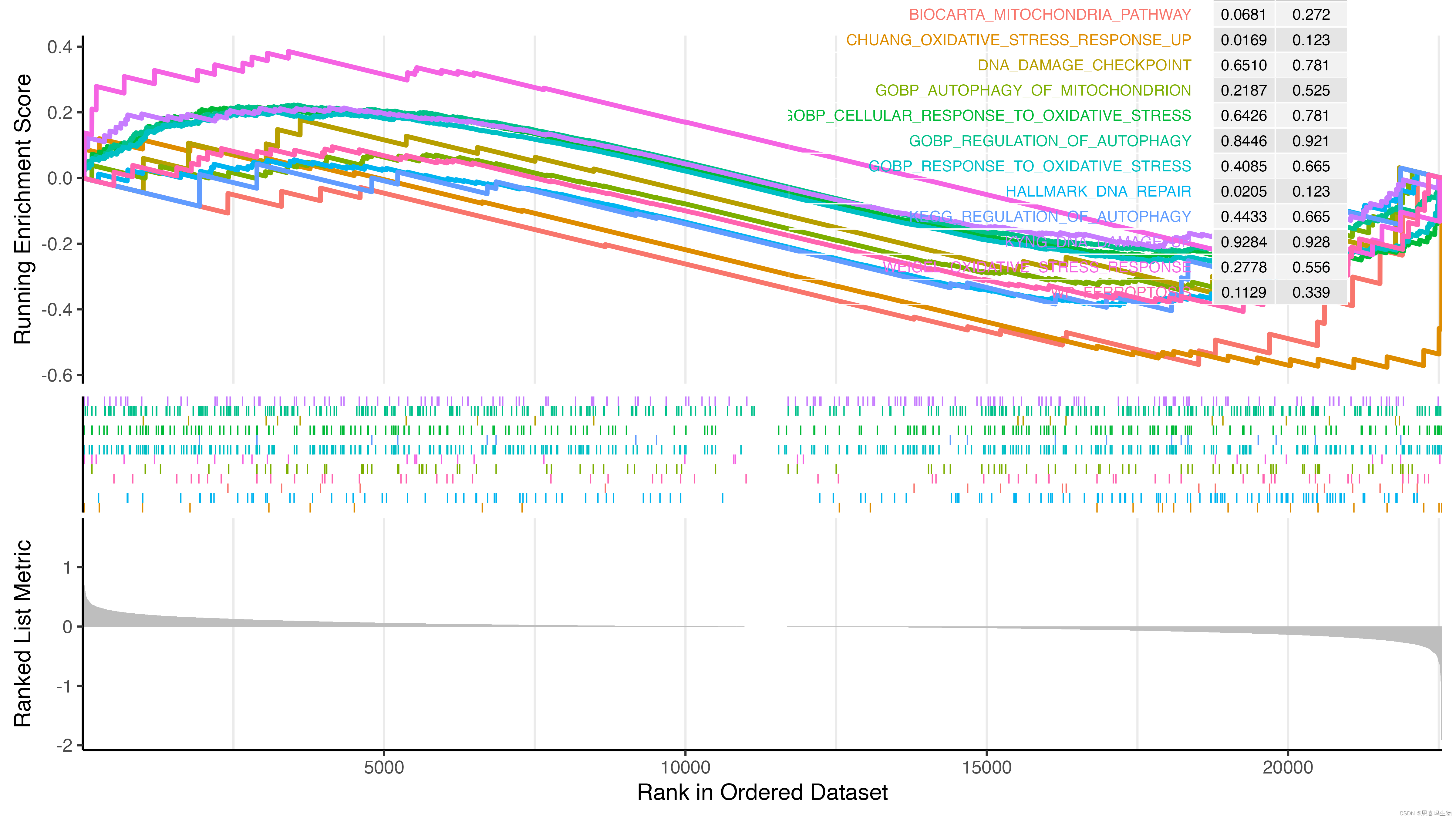
直接使用clusterProfiler::gseaplot2绘图会出现下边的结果,导致四周显示不全,线的粗细也没办法调整,因为返回的是一个aplot包中的gglist对象,没太多研究。
p1 <- clusterProfiler::gseaplot2(gsea_result, gsea_result$ID, pvalue_table = T, base_size = 18, ES_geom='line')

自定义gseaplot3函数增加了size参数调整线的粗细,也调整了margin四周边距,可以在下边gseaplot3函数的基础上继续调整,如果需要的话。
p2 <- gseaplot3(gsea_result, gsea_result$ID, pvalue_table = T, base_size = 18, ES_geom='line', size=1.8)
gseaplot3 <- function (x, geneSetID, title = "", color = "green", base_size = 15, size=1.8,rel_heights = c(1.5, 0.5, 1), subplots = 1:3, pvalue_table = FALSE, ES_geom = "line")
{library(grid)library(DOSE)gseaScores <- getFromNamespace("gseaScores", "DOSE")ES_geom <- match.arg(ES_geom, c("line", "dot"))geneList <- position <- NULLif (length(geneSetID) == 1) {gsdata <- gsInfo(x, geneSetID)}else {gsdata <- do.call(rbind, lapply(geneSetID, gsInfo, object = x))}p <- ggplot(gsdata, aes_(x = ~x)) + xlab(NULL) + theme_classic(base_size) + theme(panel.grid.major = element_line(colour = "grey92"), panel.grid.minor = element_line(colour = "grey92"), panel.grid.major.y = element_blank(), panel.grid.minor.y = element_blank()) + scale_x_continuous(expand = c(0, 0))if (ES_geom == "line") {es_layer <- geom_line(aes_(y = ~runningScore, color = ~Description), size = size)}else {es_layer <- geom_point(aes_(y = ~runningScore, color = ~Description), size = size, data = subset(gsdata, position == 1))}p.res <- p + es_layer + theme(legend.position = c(0.8, 0.8), legend.title = element_blank(), legend.background = element_rect(fill = "transparent"))p.res <- p.res + ylab("Running Enrichment Score") + theme(axis.text.x = element_blank(), axis.ticks.x = element_blank(), axis.line.x = element_blank(), plot.margin = margin(t = 0.2, r = 0.2, b = 0, l = 0.2, unit = "cm"))i <- 0for (term in unique(gsdata$Description)) {idx <- which(gsdata$ymin != 0 & gsdata$Description == term)gsdata[idx, "ymin"] <- igsdata[idx, "ymax"] <- i + 1i <- i + 1}p2 <- ggplot(gsdata, aes_(x = ~x)) + geom_linerange(aes_(ymin = ~ymin, ymax = ~ymax, color = ~Description)) + xlab(NULL) + ylab(NULL) + theme_classic(base_size) + theme(legend.position = "none", plot.margin = margin(t = -0.1, b = 0, unit = "cm"), axis.ticks = element_blank(), axis.text = element_blank(), axis.line.x = element_blank()) + scale_x_continuous(expand = c(0, 0)) + scale_y_continuous(expand = c(0, 0))if (length(geneSetID) == 1) {v <- seq(1, sum(gsdata$position), length.out = 9)inv <- findInterval(rev(cumsum(gsdata$position)), v)if (min(inv) == 0) inv <- inv + 1col <- c(rev(brewer.pal(5, "Blues")), brewer.pal(5, "Reds"))ymin <- min(p2$data$ymin)yy <- max(p2$data$ymax - p2$data$ymin) * 0.3xmin <- which(!duplicated(inv))xmax <- xmin + as.numeric(table(inv)[as.character(unique(inv))])d <- data.frame(ymin = ymin, ymax = yy, xmin = xmin, xmax = xmax, col = col[unique(inv)])p2 <- p2 + geom_rect(aes_(xmin = ~xmin, xmax = ~xmax, ymin = ~ymin, ymax = ~ymax, fill = ~I(col)), data = d, alpha = 0.9, inherit.aes = FALSE)}df2 <- p$datadf2$y <- p$data$geneList[df2$x]p.pos <- p + geom_segment(data = df2, aes_(x = ~x, xend = ~x, y = ~y, yend = 0), color = "grey")p.pos <- p.pos + ylab("Ranked List Metric") + xlab("Rank in Ordered Dataset") + theme(plot.margin = margin(t = -0.1, r = 0.2, b = 0.2, l = 0.2, unit = "cm"))if (!is.null(title) && !is.na(title) && title != "") p.res <- p.res + ggtitle(title)if (length(color) == length(geneSetID)) {p.res <- p.res + scale_color_manual(values = color)if (length(color) == 1) {p.res <- p.res + theme(legend.position = "none")p2 <- p2 + scale_color_manual(values = "black")}else {p2 <- p2 + scale_color_manual(values = color) + theme(legend.position = "none",plot.margin=margin(t = 0.1, r = 0.2, b = 0.1, l = 0.2, unit = "cm"))}}if (pvalue_table) {pd <- x[geneSetID, c("Description", "pvalue", "p.adjust")]rownames(pd) <- pd$Descriptionpd <- pd[, -1]for (i in seq_len(ncol(pd))) {pd[, i] <- format(pd[, i], digits = 3)}tp <- tableGrob2(pd, p.res)p.res <- p.res + theme(legend.position = "none",plot.margin=margin(t = 0.8, r = 0.2, b = 0.2, l = 0.2,unit = "cm")) + annotation_custom(tp, xmin = quantile(p.res$data$x, 0.5), xmax = quantile(p.res$data$x, 0.95), ymin = quantile(p.res$data$runningScore, 0.75), ymax = quantile(p.res$data$runningScore, 0.9))}plotlist <- list(p.res, p2, p.pos)[subplots]n <- length(plotlist)plotlist[[n]] <- plotlist[[n]] + theme(axis.line.x = element_line(), axis.ticks.x = element_line(), axis.text.x = element_text())if (length(subplots) == 1) return(plotlist[[1]] + theme(plot.margin = margin(t = 0.2, r = 0.2, b = 0.2, l = 0.2, unit = "cm")))if (length(rel_heights) > length(subplots)) rel_heights <- rel_heights[subplots]aplot::gglist(gglist = plotlist, ncol = 1, heights = rel_heights)}gsInfo <- function(object, geneSetID) {geneList <- object@geneListif (is.numeric(geneSetID))geneSetID <- object@result[geneSetID, "ID"]geneSet <- object@geneSets[[geneSetID]]exponent <- object@params[["exponent"]]df <- gseaScores(geneList, geneSet, exponent, fortify=TRUE)df$ymin <- 0df$ymax <- 0pos <- df$position == 1h <- diff(range(df$runningScore))/20df$ymin[pos] <- -hdf$ymax[pos] <- hdf$geneList <- geneListdf$Description <- object@result[geneSetID, "Description"]return(df)
}tableGrob2 <- function(d, p = NULL) {# has_package("gridExtra")d <- d[order(rownames(d)),]tp <- gridExtra::tableGrob(d)if (is.null(p)) {return(tp)}# Fix bug: The 'group' order of lines and dots/path is differentp_data <- ggplot_build(p)$data[[1]]# pcol <- unique(ggplot_build(p)$data[[1]][["colour"]])p_data <- p_data[order(p_data[["group"]]), ]pcol <- unique(p_data[["colour"]])## This is fine too## pcol <- unique(p_data[["colour"]])[unique(p_data[["group"]])] j <- which(tp$layout$name == "rowhead-fg")for (i in seq_along(pcol)) {tp$grobs[j][[i+1]][["gp"]] <- gpar(col = pcol[i])}return(tp)
}






)





外星语词典全过程文档及程序)







