微生物 研究
Background
背景
While a New York Subway station is bustling with swarms of businessmen, students, artists, and millions of other city-goers every day, its floors, railings, stairways, toilets, walls, kiosks, and benches are teeming with non-human life. The microbial ecosystem, or the complex web of relationships that microorganisms have with one another and with the environment, is omnipresent in shared public transportation spaces. The PathoMap, a 2013 project led by Cornell Professor Christopher Mason, was the first of an annual series of DNA collection projects at various locations around the world. A sampling from NYC subways found that only half of the DNA matched known organisms. This initial project confirmed that the urban microbiome was still a relatively unexplored field, virtually begging for researchers to seize the opportunity. The success of the project yielded the creation of the MetaSUB international consortium, and ever since, intensive studies have been carried out with microbial samples from urban locations around the globe. In addition to being a relatively new field of work, the applications of such projects are endless.
纽约地铁站每天都挤满了大批商人,学生,艺术家和数百万其他城市居民,但它的地板,栏杆,楼梯,厕所,墙壁,信息亭和长椅上充斥着非人类的生活。 微生物生态系统,或微生物彼此之间以及与环境之间复杂的关系网,在共享的公共交通空间中无处不在。 由康奈尔大学教授克里斯托弗·梅森(Christopher Mason)领导的2013年PathoMap项目是全球各地每年一次的DNA收集项目的第一个系列。 从纽约地铁的一个样本中发现,只有一半的DNA与已知生物匹配。 该项目初始确认该城市的微生物仍然是一个相对未开发领域,几乎乞求的研究人员抓住机会。 该项目的成功促成了MetaSUB国际财团的创立,从那时起,就对来自全球城市地区的微生物样本进行了深入研究。 除了是一个相对较新的工作领域之外,此类项目的应用也无穷无尽。
But what happens after the samples are collected? As I learned this summer, there is no magical one-step formula that outputs clean, categorized, analyzed, and graphed data. I had the opportunity to work with MetaSUB microbial data and learn the painstaking yet satisfying process of biological data manipulation and visualization.
但是收集样本后会怎样? 正如我今年夏天了解到的那样,没有神奇的一步式公式可以输出干净,分类,分析和图形化的数据。 我有机会处理MetaSUB微生物数据,并学习了艰苦而又令人满意的生物数据处理和可视化过程。
Collecting & Cleaning the Data in Linux Bash Terminal
在Linux Bash Terminal中收集和清理数据
Before the analysis is run, swab samples are collected from specified locations, and DNA libraries are prepared for paired-end sequencing. Since both ends of the fragment are sequenced, this type of sequencing allows for more precise reading of the DNA, results in better alignment, and detects any rearrangements.
在运行分析之前,应从指定的位置收集拭子样品,并准备DNA文库用于双末端测序。 由于片段的两端均已测序,因此这种类型的测序可更精确地读取DNA,更好的比对并检测任何重排。
While the biological sampling yields a plethora of data, not all of it is relevant for analysis. Adapter sequences, low-quality bases, and human DNA were all extraneous data points in the set. For my project, the first goal was to clean and categorize this data in the Linux Bash Terminal to run visual analyses in R Studio.
尽管生物采样产生了大量数据,但并非所有数据都与分析相关。 接头序列,低质量碱基和人类DNA都是该集中的无关数据点。 对于我的项目,首要目标是在Linux Bash Terminal中清理和分类这些数据,以便在R Studio中运行可视化分析。
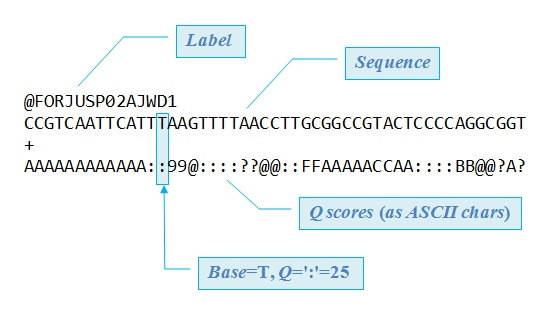
The Linux Bash Terminal is used for all reading and cleaning of the data. Read files are stored in a FASTQ format (as seen in Figure 1), and the following is the structure of this file type:
Linux Bash Terminal用于所有读取和清除数据。 读取文件以FASTQ格式存储(如图1所示),以下是该文件类型的结构:
- Line 1 of the file contains the identifier of the sequence which summarizes where the sequence of bases is found 文件的第1行包含序列的标识符,该标识符概括了在何处发现碱基序列
- Line 2 provides the actual raw letters of the nitrogenous base sequence 第2行提供了含氮碱基序列的实际原始字母
- Line 3 starts with the plus sign and may be followed by the sequence identifier again 第3行以加号开头,并且可能再次由序列标识符跟随
- Line 4 is the last line of the FASTQ file, and it contains the quality score of each base in the format of an ASCII symbol, each symbol corresponding to a Phred Quality (Q) score 第4行是FASTQ文件的最后一行,它包含ASCII符号格式的每个基准的质量得分,每个符号对应于Phred Quality(Q)得分
However, the raw files still have unwanted DNA fragments, including adapters (which are used to ligate the DNA molecules) and low-quality bases (which can be identified using the ASCII or Phred Quality scores). The Linux AdapterRemoval function is used to remove these adapter sequences and trim the data of low-quality bases, as seen in Figure 2.
但是,原始文件仍然具有不需要的DNA片段,包括衔接子(用于连接DNA分子)和低质量的碱基(可以使用ASCII或Phred Quality分数进行标识)。 Linux AdapterRemoval函数用于删除这些适配器序列并修剪低质量基数的数据,如图2所示。
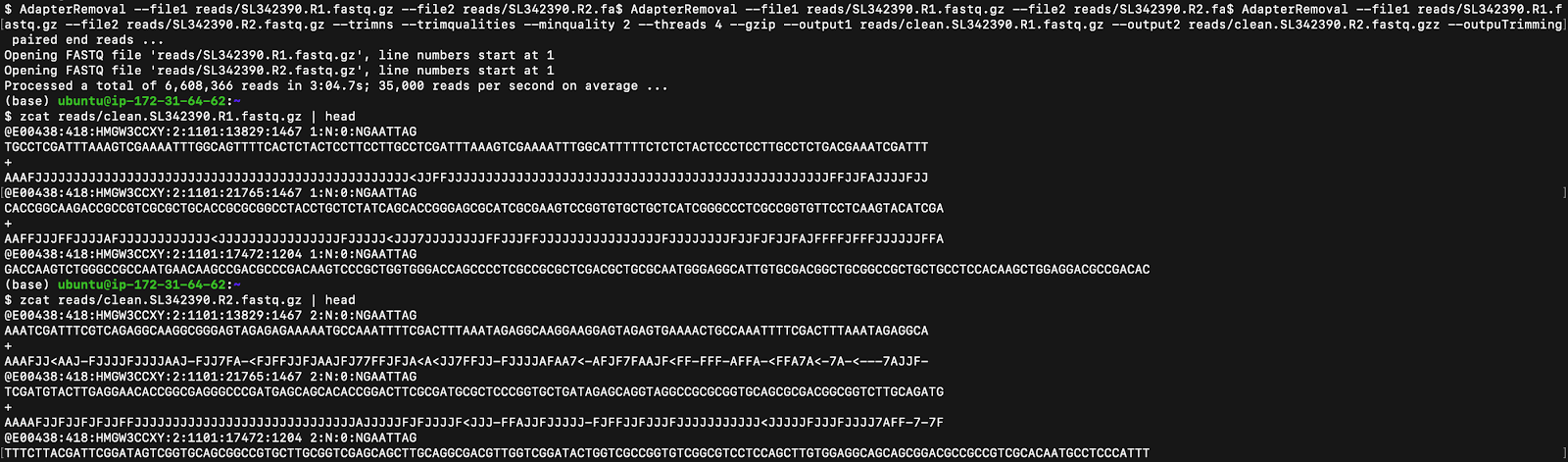
New FASTQ files are created from the trimmed reads after the sample reads are identified, the Adapter Removal function is used, the reads are filtered with a minimum quality score, and the files are unzipped as seen in the first line of code in Figure 2.
在识别样本读取后,使用修剪后的读取创建新的FASTQ文件,使用适配器移除功能,以最低质量得分过滤读取,并解压缩文件,如图2中第一行代码所示。
Although these sequences have been removed, the reads are still not thoroughly cleaned. For this project, only microbial data is needed. However, DNA samples from a public transportation space will contain mostly (close to 99%) human DNA since the bacterial genome is more than 1600 times smaller than the human one. Fortunately, most of this human DNA has already been stripped in earlier stages of this project, though some may remain. The bowtie2 function in Linux must be used to completely rid the reads of human DNA.
尽管已删除了这些序列,但仍未彻底清除读段。 对于此项目,仅需要微生物数据。 但是,来自公共交通场所的DNA样本将主要(接近99%)包含人类DNA,因为细菌基因组的大小是人类基因组的1600倍以上。 幸运的是,大多数人类DNA在该项目的早期阶段已经被剥离,尽管可能还会保留一些。 Linux中的bowtie2函数必须用于完全清除人类DNA的读码。
The bowtie2 function works in a fairly intuitive way. Rather than going through each file and identifying whether or not there is human DNA (which would yield a terrible run time), the algorithm utilizes a human index reference genome. Many human reference genomes have been created for such purposes, and the most commonly used one is reference genome 38, which is used here. To provide an analogy for how the function works, the way I thought of this was like a book — to find a term or specific sentence in a book, it is far easier to check the index to see what page it is on rather than flipping through the full text. Likewise, suppose we provide bowtie with the information on the human reference genome. In that case, it will be much more efficient because the program will know where to go on the genome since the reference index narrows it down. From the function’s output in Figure 3, it can be seen that 0.95% of the genome aligned with human DNA, meaning that the reads were already thoroughly cleaned, but not yet entirely stripped as it is now. New files were created that had cleaned reads.
bowtie2函数以相当直观的方式工作。 该算法不是遍历每个文件并识别是否存在人类DNA(这将导致可怕的运行时间),而是利用人类索引参考基因组。 为此目的已经创建了许多人类参考基因组,最常用的是参考基因组38,在此使用。 为了给函数的工作方式提供一个类比,我想到的方式就像一本书—在书中查找一个术语或特定句子,检查索引以查看它所在的页面比翻页要容易得多。通过全文。 同样,假设我们向领结提供有关人类参考基因组的信息。 在这种情况下,它将更有效率,因为程序会知道基因组的位置,因为参考索引会缩小基因组的范围。 从图3的函数输出中可以看出,基因组的0.95%与人类DNA对齐,这意味着读取的片段已被彻底清洗,但尚未像现在一样被完全剥离。 创建的新文件已清除读取。
After cleaning the data of unwanted reads (which could have meant either adapter sequences, low-quality bases, or human DNA), it is essential to identify the amount of data we will be using. To count the total number of reads (and we will count them at each stage of the process up until now) the zcat function is used to calculate the total number of FASTQ file lines and the output is divided by four since all reads are spread across the four lines of each FASTQ file.
清除不需要的读数(可能意味着衔接子序列,低质量的碱基或人类DNA)的数据后,确定我们将要使用的数据量至关重要。 要计算读取的总数(到目前为止,我们将在过程的每个阶段进行计数),zcat函数用于计算FASTQ文件行的总数,并且由于所有读取分散在输出中,因此输出结果被四分每个FASTQ文件的四行。
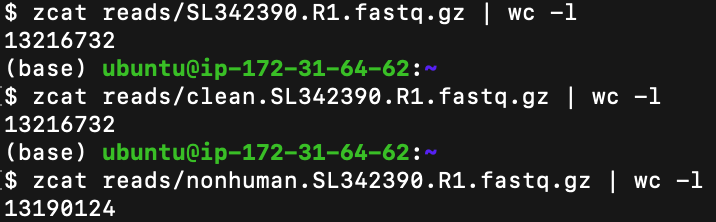
- Initial # of reads: 3,304,183 reads 初始读取次数:3,304,183次读取
- # of reads after AdapterRemoval: 3,304,183 适配器卸下后的读取次数:3,304,183
- # of reads after Bowtie 2 Alignment: 3,297,531 Bowtie 2对齐后的读取次数:3,297,531
Furthermore, to learn more about the data, the GC (guanine-cytosine percent) content is calculated using the qc-stats function, as seen in Figure 5.
此外,要了解有关数据的更多信息,请使用qc-stats函数计算GC(鸟嘌呤-胞嘧啶百分比)含量,如图5所示。
The significance of the GC content is that there is a correlation between GC content and types of bacteria. Bacteria that might be specific to a particular biome may have comparatively lower or higher GC contents than those of other biomes. There is also a strong correlation between GC content and the stability of DNA, since G-C base pairs have one more hydrogen bond than A-T base pairs. Therefore, a higher GC content means higher required temperature for denaturing the DNA, and this information is useful for PCR amplification and identifying the source or environment of the bacteria. Next, using a program called Kraken, the taxonomically categorized data can be printed in the Linux Terminal, as seen in Figure 6.
GC含量的意义在于GC含量和细菌类型之间存在相关性。 特定于特定生物群落的细菌可能具有比其他生物群落相对较低或较高的GC含量。 GC含量与DNA稳定性之间也有很强的相关性,因为GC碱基对比AT碱基对具有更多的氢键。 因此,较高的GC含量意味着使DNA变性所需的较高温度,并且此信息可用于PCR扩增和鉴定细菌的来源或环境。 接下来,使用称为Kraken的程序,可以在Linux Terminal中打印分类分类的数据,如图6所示。
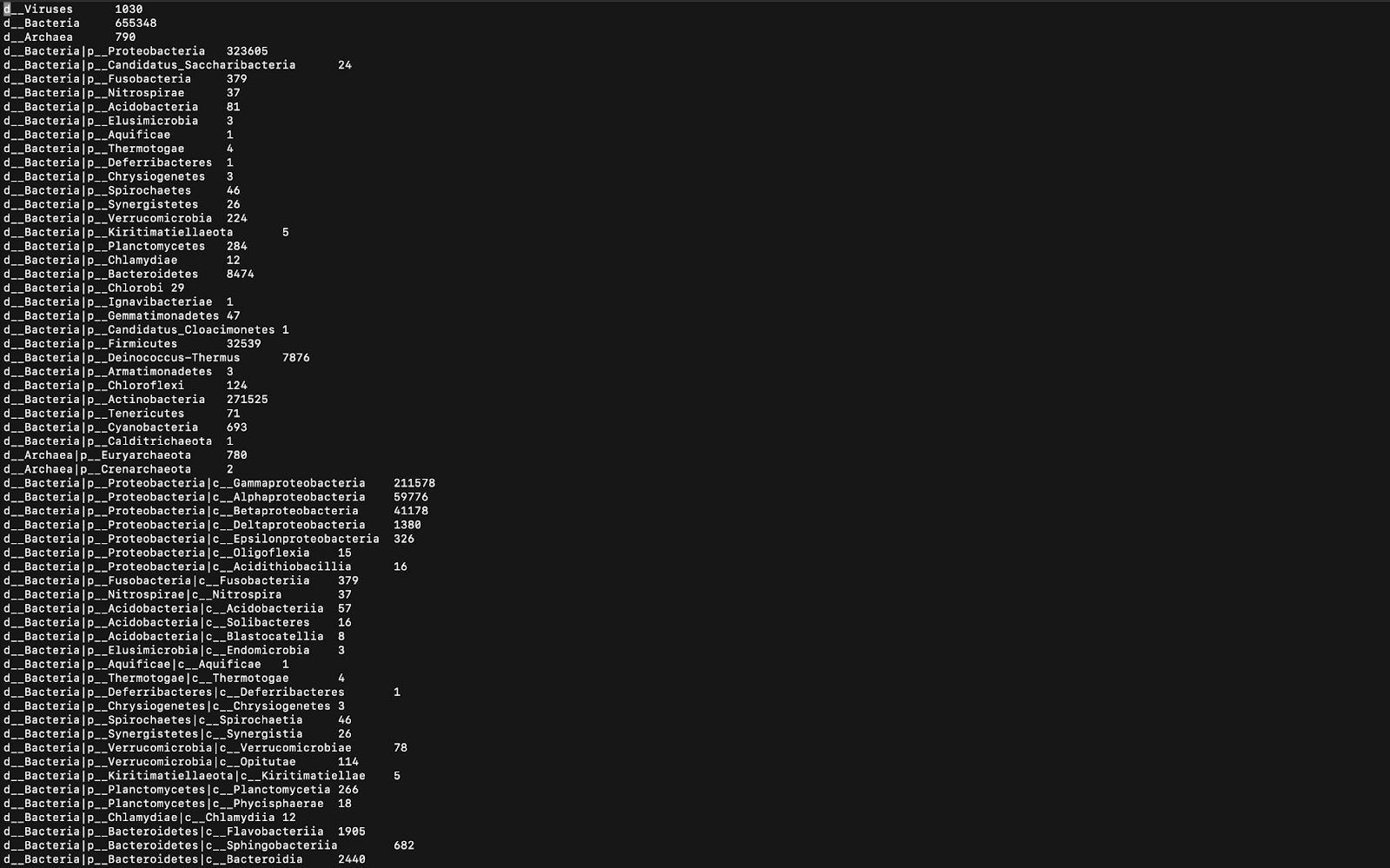
Kraken’s output is handy, as the actual classifications of the microbes can now be seen. In Figure 6, only a minimal number of the total classifications can be seen (that’s all that fit in the screenshot). The letters before the rankings are all abbreviations for Domain, Kingdom, Phylum, Class, Order, Family, Genus, and Species (the taxonomy order). The numbers at the end of each line represent the proportion of the corresponding species (or domain, phylum, etc. if the species was not identified) in the sample. The program is not able to identify many of the species because those are yet to be discovered.
Kraken的输出非常方便,因为现在可以看到微生物的实际分类。 在图6中,只能看到最小数量的总分类(这就是屏幕截图中的所有内容)。 排名前的字母都是Domain,Kingdom,Phylum,Class,Order,Family,Genus和Species(分类法顺序)的缩写。 每行末尾的数字代表样本中相应物种(或领域,门类等,如果未识别出)的比例。 该程序无法识别许多物种,因为尚未发现这些物种。
Visual & Categorical Analysis in R
R中的视觉和分类分析
After finally cleaning and classifying the raw data, we will move to R to visualize and categorize the data. Mystery DNA will be added to the dataset to simplify the purpose of using R, and our new goal will be to identify this data through visualization.
在最终清理和分类原始数据之后,我们将移至R以对数据进行可视化和分类。 神秘DNA将添加到数据集中,以简化使用R的目的,而我们的新目标将是通过可视化识别此数据。
The first step in R Studio is to call the packages needed to visualize the data — ggplot2 is a package used to create all sorts of graphs and plots. The data is then read in using the read.csv function. Next, the geom_bar function of ggplot2 is used to create a barplot of the phylum classifications of the various samples:
R Studio的第一步是调用可视化数据所需的软件包-ggplot2是用于创建各种图形和绘图的软件包。 然后使用read.csv函数读取数据。 接下来,使用ggplot2的geom_bar函数创建各种样本的门类分类的barplot:
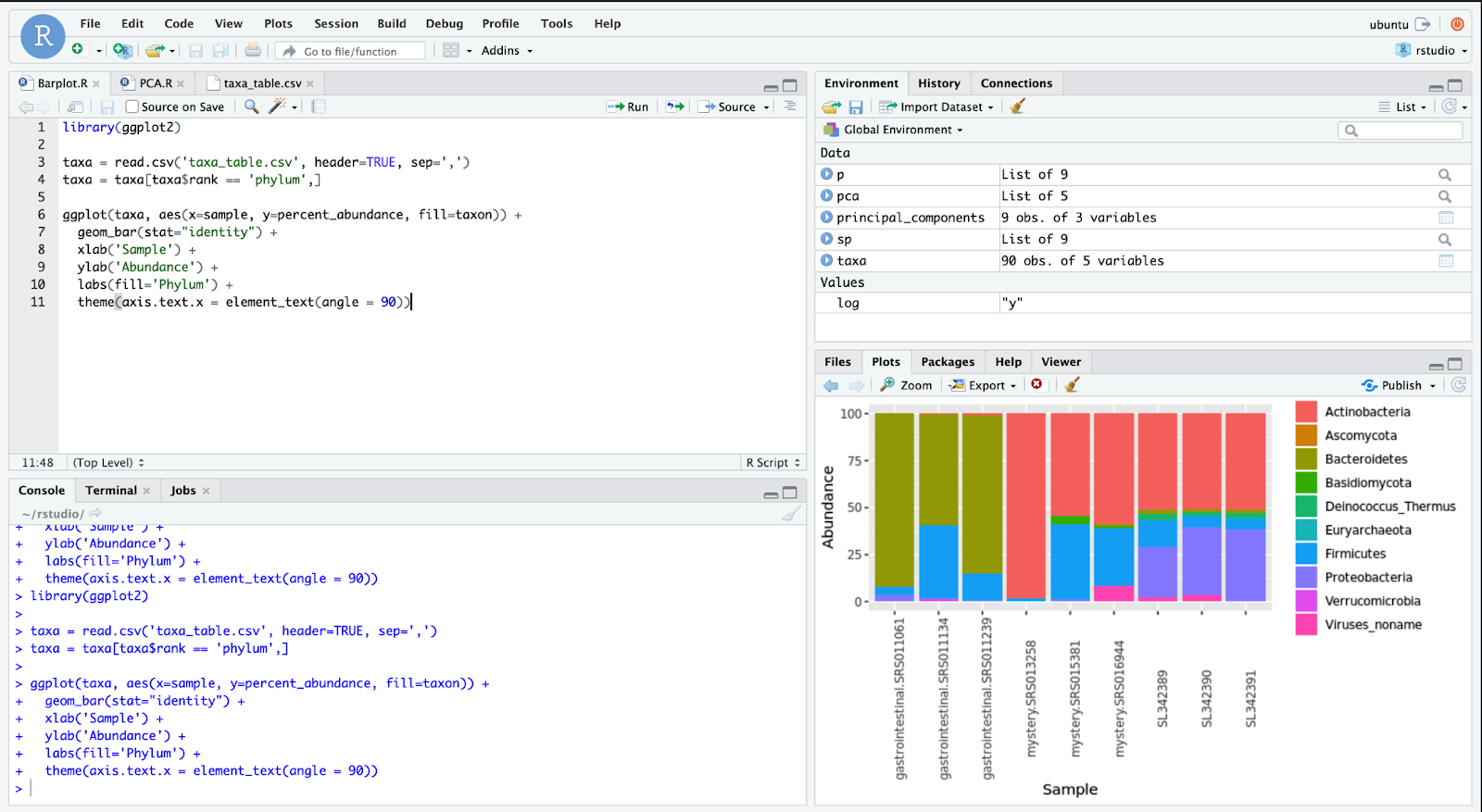
As shown in the bar graph from Figure 7, there are three samples from each environment — the first three are gastrointestinal data, then there are three samples from a mystery environment, and the final three come from shared transportation spaces. Already, some distinctions are apparent between the three groups. Gastrointestinal samples contain far more Bacteroidetes than the two other environments. The first mystery sample almost entirely consists of Actinobacteria. Despite these initial observations, it is difficult to come to a conclusion on the mystery samples since they are only grouped broadly by the microbes’ phyla, as seen in line 4 of the code. But before we make a bar graph of more specific taxonomic groups, it is necessary to analyze the variance among the samples. Principal Component Analysis (PCA), which decreases the number of variables or dimensions in the dataset, makes it easier to see the variance and patterns among groups in the dataset. As seen in Figure 8, some lines of code are added to create this PCA plot, and the points are colored according to their environment.
如图7的条形图所示,每个环境有三个样本-前三个样本是胃肠道数据,然后三个样本来自一个神秘环境,最后三个样本来自共享运输空间。 这三组之间已经有一些明显的区别。 胃肠道样品中的拟杆菌含量远高于其他两种环境。 第一个神秘样本几乎完全由放线菌组成。 尽管有这些最初的观察结果,但很难对神秘样本做出结论,因为如代码第4行所示,这些样本仅按微生物的门类进行了大致分组。 但是在制作更具体的分类组的条形图之前,有必要分析样本之间的差异。 主成分分析(PCA)减少了数据集中变量或维的数量,使查看数据集中各组之间的差异和模式变得更加容易。 如图8所示,添加了一些代码行以创建此PCA图,并且根据其环境为这些点着色。
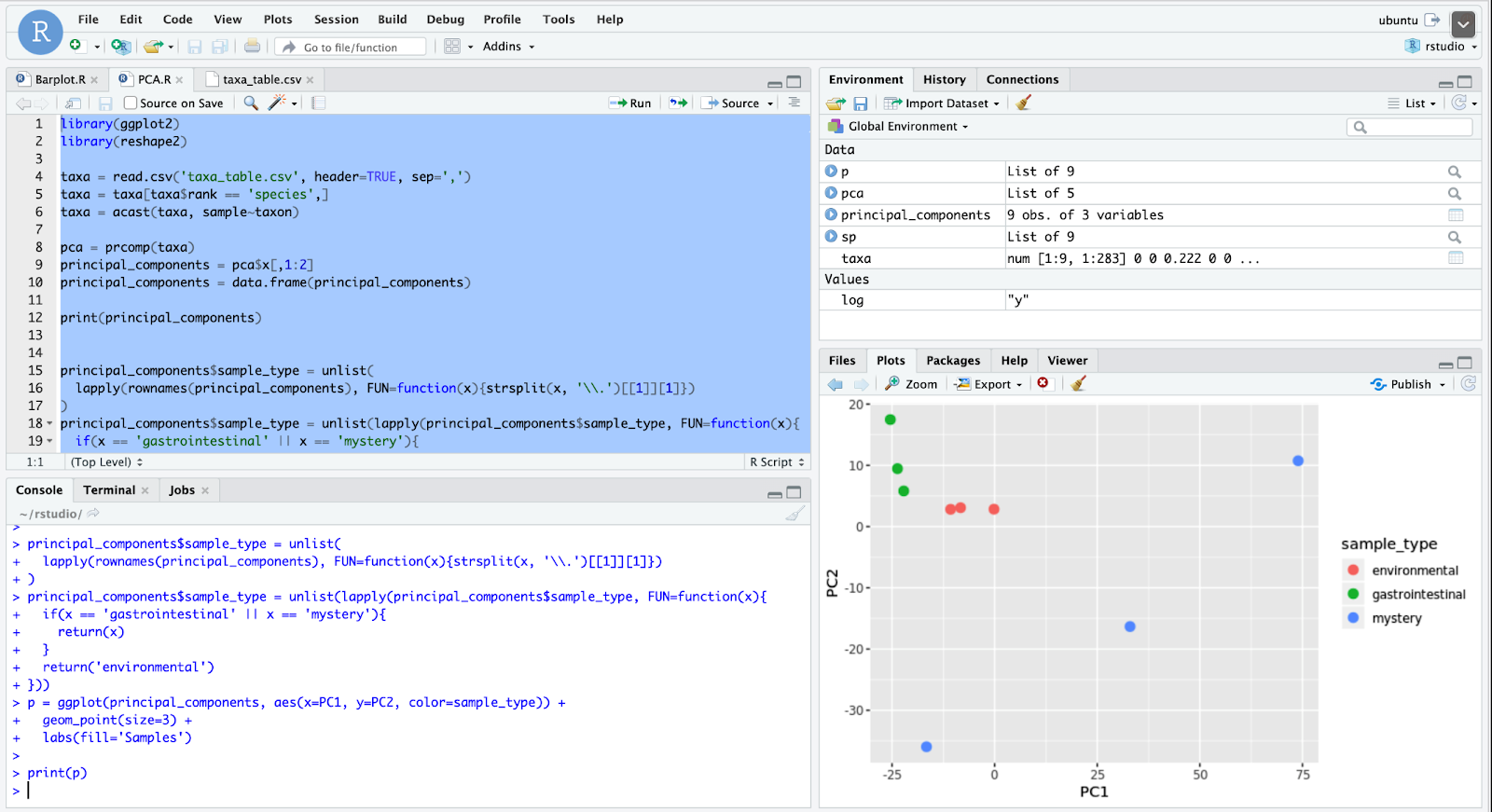
A few conclusions can be drawn from the plot in Figure 8. One is that both the environmental (mass transit) and gut samples lack variance since points are clustered tightly within each group. Meanwhile, mystery samples are not nearly as correlated since their points are spread apart, which tells us that those samples come from an environment where the types of microbes vary. From the initial conclusions, one might be quick to suggest that the environment is one of soil, as this environment is known to contain large amounts of actinobacteria and is also very variable. However, this conclusion is likely flawed since a more specific grouping of the first bar graph is needed to compare to other studies. When ranked by genus, the taxonomic abundances of the mystery samples look far more similar to the one on the right, which comes from a research paper on skin pore taxonomic abundance, not soil. Even though Figure 9 comes from a study on the impact of pomegranate juice on human skin microbiota, we can compare its control groups with our data as well.
从图8中的曲线可以得出一些结论。一个是环境(大众运输)样本和肠道样本都缺乏方差,因为每个组中的点紧密聚集。 同时,由于神秘样本的点分散开,因此它们之间的相关度也不高,这告诉我们这些样本来自微生物种类不同的环境。 从最初的结论来看,可能会很快暗示环境是土壤的一种,因为已知该环境包含大量放线菌,而且变化很大。 但是,该结论可能有缺陷,因为需要将第一条形图的更具体分组与其他研究进行比较。 当按属进行排序时,神秘样本的生物分类丰度看起来与右侧样本更加相似,后者来自有关皮肤 毛Kong生物分类丰度而不是土壤的研究论文。 即使图9来自石榴汁对人类皮肤微生物群影响的研究,我们也可以将其对照组与我们的数据进行比较。
As it turns out, Propionibacterium and Staphylococcus were the most common microbes in the mystery samples. Still, we would lack this specificity without narrowing down the taxonomic grouping from phylum to genus. Indeed, human skin pores were the correct environment from which the mystery samples were taken.
事实证明,丙酸杆菌和葡萄球菌是神秘样品中最常见的微生物。 但是,如果不缩小分类范围,从门类到属类,我们将缺乏这种特异性。 确实,人的皮肤毛Kong是从中取样神秘样品的正确环境。
Conclusions & Significance
结论与意义
Although the main visualization and analysis stages of this project were simplified compared to the actual processes of MetaSUB, projects like these bring light to the significance of their research. With so many unknown species lurking in mass transit spaces around the world, studying the microbiome can spur rapid innovation in the field. The possibilities of the research applications are endless — the data can play a crucial role in urban planning, city design, public health, and the discovery of new species. As the MetaSUB website says, the “data will… [enable] an era of more quantified, responsive, and smarter cities.” Especially with this past year’s COVID19 pandemic, researching and analyzing the microbial environments of spaces that billions of people share is all the more important, if not necessary.
尽管与MetaSUB的实际过程相比,该项目的主要可视化和分析阶段得到了简化,但类似的项目使他们的研究意义更为明显。 由于世界各地的大众运输空间中潜伏着如此众多的未知物种,因此研究微生物组可以促进该领域的快速创新。 研究应用的可能性是无限的-数据可以在城市规划,城市设计,公共卫生和新物种发现中发挥关键作用。 正如MetaSUB网站所说,“数据将……(使)一个更量化,响应更快和更智能的城市时代”。 尤其是在去年的COVID19大流行中,研究和分析数十亿人共享的空间的微生物环境,即使不是必须的话,也变得尤为重要。
Credits
学分
MILRD for providing the data, steps for the process, and invaluable mentorship.
MILRD用于提供数据,流程步骤和宝贵的指导。
Sources Used
资料来源
http://metasub.org/
http://metasub.org/
https://www.researchgate.net/figure/Relative-abundance-of-skin-microbiota-before-and-after-pomegranate-and-placebo_fig1_336383548
https://www.researchgate.net/figure/Relative-abundance-of-skin-microbiota-before-and-after-pomegranate-and-placebo_fig1_336383548
翻译自: https://medium.com/swlh/microbial-surveillance-how-it-works-why-its-important-880f67aaa8b0
微生物 研究
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。如若转载,请注明出处:http://www.mzph.cn/news/389314.shtml
如若内容造成侵权/违法违规/事实不符,请联系多彩编程网进行投诉反馈email:809451989@qq.com,一经查实,立即删除!






思想及实现)











