目录
- 前言
- 代码示例
- 1、寻找函数最大值
- 2、句子匹配
前言
遗传算法就是在一个解空间上,随机的给定一组解,这组解称为父亲种群,通过这组解的交叉,变异,构建出新的解,称为下一代种群,然后在目前已有的所有解中抽取表现好的解组成新的父亲种群,然后继续上面的过程,直到达到了迭代条件或者获取到了最优解。
进化算法流程框架
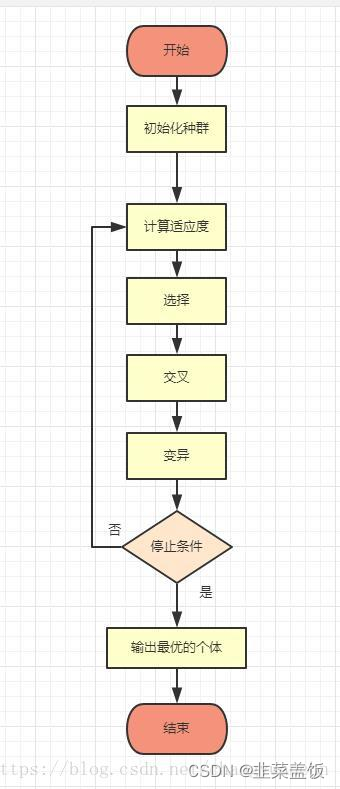
下面我们来解释下这个流程图里面的一些概念
- 适应度
所谓的适应度,本质上可以理解为一个代价函数,或者一个规则,通过对初始种群中的个体计算适应度,能够得到对初始种群中的个体是否优劣的一个度量
- 选择
选择操作是根据种群中的个体的适应度函数值所度量的优、劣程度决定它在下一代是被淘汰还是被遗传。
- 交叉
交叉操作是将选择的两个个体p1和 p2
作为父母节点,将两者的部分码值进行交换。假设有下面的两个节点的二进制编码表示:
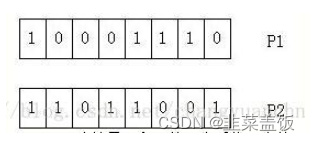
随机产生一个1到7之间的随机数,假设为3,则将p1和 p2的低三位进行互换,如下图所示,就完成了交叉操作:
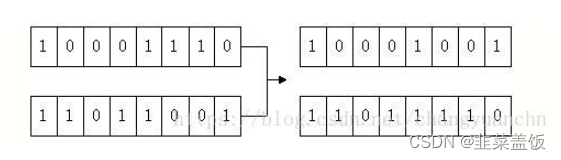
- 变异
变异操作就是改变一个DNA序列上某一个点,

我们以一定的概率进行变异的操作,例如上方的DNA序列的第6个结点,由1变成了0

代码示例
1、寻找函数最大值

首先我们生成POP_SIZE个长度为DNA_SIZE的DNA序列

pop = np.random.randint(2, size=(POP_SIZE, DNA_SIZE))
定义函数图像
def F(x):"""定义一个函数,就是图像中的线条:param x::return:"""return np.sin(10*x)*x + np.cos(2*x)*x # to find the maximum of this function
我们可以将DNA转换为函数中x轴中的某个点,将0 1 DNA序列翻译成范围在(0, 5)的数字
def translateDNA(pop):"""将0 1 DNA序列翻译成范围在(0, 5)的数字:param pop::return:"""return pop.dot(2 ** np.arange(DNA_SIZE)[::-1]) / float(2**DNA_SIZE-1) * X_BOUND[1]
x轴上对应的y轴上的值,我们将其称为适应度
def get_fitness(pred):"""获取个体的适用度:param pred::return:"""return pred + 1e-3 - np.min(pred) #防止适应度为负数
我们从种群中选择个体
注意: 并不是每次都选择适应度最高的个体,只是适应度高的个体选择的概率比较大,其它适应度较低的个体,选择的概率小,将来适应度低的个体变异后,适应度可能会变大
def select(pop, fitness): # nature selection wrt pop's fitness"""从种群中选择:param pop::param fitness::return: 所选个体的下标,可重复选择replace=True"""idx = np.random.choice(np.arange(POP_SIZE), size=POP_SIZE, replace=True,p=fitness/fitness.sum())return pop[idx]pop = select(pop, fitness)
之后我们对选择后的种群进行交叉和变异
def crossover(parent, pop): # mating process (genes crossover)"""交叉配对:param parent::param pop::return:"""if np.random.rand() < CROSS_RATE:i_ = np.random.randint(0, POP_SIZE, size=1) # select another individual from pop 从种群中选择一个个体,cross_points = np.random.randint(0, 2, size=DNA_SIZE).astype(np.bool_) # choose crossover points 生成要交叉的位置parent[cross_points] = pop[i_, cross_points] # mating and produce one child 进行交叉return parentdef mutate(child):"""变异:param child::return:"""for point in range(DNA_SIZE):if np.random.rand() < MUTATION_RATE:child[point] = 1 if child[point] == 0 else 0return child# 复制一个副本pop_copy = pop.copy()# 进行遗传 和变异for parent in pop:child = crossover(parent, pop_copy)child = mutate(child)parent[:] = child # parent is replaced by its child 父亲个体被孩子代替
最后,我们对其迭代多次
for _ in range(N_GENERATIONS):# 得到图像上的y值F_values = F(translateDNA(pop))# 更新散点图if 'sca' in globals(): sca.remove()sca = plt.scatter(translateDNA(pop), F_values, s=200, lw=0, c='red', alpha=0.5)plt.pause(0.1)# 获取每一个个体的适应度fitness = get_fitness(F_values)#获取最好适用度的个体下标i = np.argmax(fitness)print("最优DNA",pop[i,:])pop = select(pop, fitness)# 复制一个副本pop_copy = pop.copy()# 进行遗传 和变异for parent in pop:child = crossover(parent, pop_copy)child = mutate(child)parent[:] = child # parent is replaced by its child 父亲个体被孩子代替
完整代码:
#!/usr/bin/env python
# -*- coding:utf-8 -*-
import timeimport numpy as np
import matplotlib.pyplot as pltDNA_SIZE = 10 # DNA length DNA长度
POP_SIZE = 100 # population size 种群大小
CROSS_RATE = 0.8 # mating probability (DNA crossover) 交叉配对的概率
MUTATION_RATE = 0.003 # mutation probability 编译的概率
N_GENERATIONS = 200 # 代数
X_BOUND = [0, 5] # x upper and lower bounds x轴范围def F(x):"""定义一个函数,就是图像中的线条:param x::return:"""return np.sin(10*x)*x + np.cos(2*x)*x # to find the maximum of this functiondef get_fitness(pred):"""获取个体的适用度:param pred::return:"""return pred + 1e-3 - np.min(pred) #防止适应度为负数
def translateDNA(pop):"""将0 1 DNA序列翻译成范围在(0, 5)的数字:param pop::return:"""return pop.dot(2 ** np.arange(DNA_SIZE)[::-1]) / float(2**DNA_SIZE-1) * X_BOUND[1]def select(pop, fitness): # nature selection wrt pop's fitness"""从种群中选择choice replace是true 那么适应度高的基因会被重复多选,达到进化的目的:param pop::param fitness::return: 所选个体的下标,可重复选择replace=True"""idx = np.random.choice(np.arange(POP_SIZE), size=POP_SIZE, replace=True,p=fitness/fitness.sum())return pop[idx]def crossover(parent, pop): # mating process (genes crossover)"""交叉配对:param parent::param pop::return:"""if np.random.rand() < CROSS_RATE:i_ = np.random.randint(0, POP_SIZE, size=1) # select another individual from pop 从种群中选择一个个体,cross_points = np.random.randint(0, 2, size=DNA_SIZE).astype(np.bool_) # choose crossover points 生成要交叉的位置parent[cross_points] = pop[i_, cross_points] # mating and produce one child 进行交叉return parentdef mutate(child):"""变异:param child::return:"""for point in range(DNA_SIZE):if np.random.rand() < MUTATION_RATE:child[point] = 1 if child[point] == 0 else 0return childif __name__ == '__main__':# initialize the pop DNA s生成0-1的POP_SIZE行 DNA_SIZE列的种群pop = np.random.randint(2, size=(POP_SIZE, DNA_SIZE))plt.ion() # something about plotting# x轴x = np.linspace(*X_BOUND, 200)plt.plot(x, F(x))# 迭代N_GENERATIONS次for _ in range(N_GENERATIONS):# 得到图像上的y值F_values = F(translateDNA(pop))# 更新散点图if 'sca' in globals(): sca.remove()sca = plt.scatter(translateDNA(pop), F_values, s=200, lw=0, c='red', alpha=0.5)plt.pause(0.1)# 获取每一个个体的适应度fitness = get_fitness(F_values)#获取最好适用度的个体下标i = np.argmax(fitness)print("最优DNA",pop[i,:])pop = select(pop, fitness)# 复制一个副本pop_copy = pop.copy()# 进行遗传 和变异for parent in pop:child = crossover(parent, pop_copy)child = mutate(child)parent[:] = child # parent is replaced by its child 父亲个体被孩子代替plt.ioff()plt.show()2、句子匹配
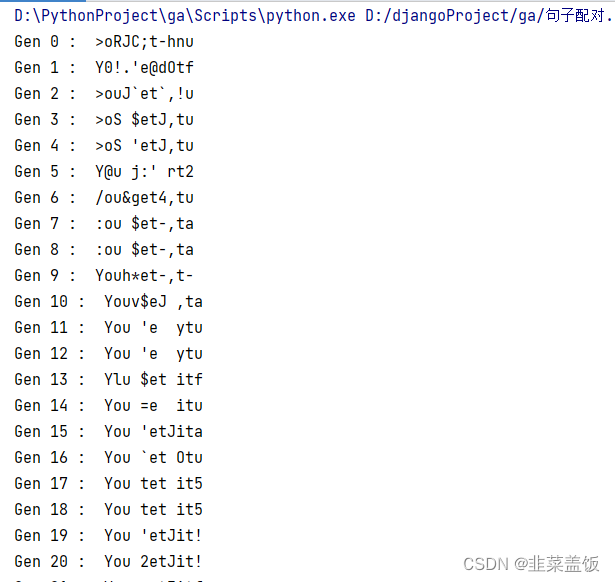
句子匹配与寻找函数最大值的区别就是DNA的表示不同,寻找函数最大值的DNA为0 1表示的,而句子匹配将字母转换为ASCII表示,多个字母组成的ASCII序列就表示为DNA
即 [ 89 111 117 32 103 101 116 32 105 116 33] 用ASCII翻译过来就是句子'You get it!'
其次 代码将进化算法封装到了一个类中
完整代码:
#!/usr/bin/env python
# -*- coding:utf-8 -*-import numpy as npTARGET_PHRASE = b'You get it!' # target DNA 目标句子
POP_SIZE = 300 # population size 种群大小
CROSS_RATE = 0.4 # mating probability (DNA crossover) 交叉的概率
MUTATION_RATE = 0.01 # mutation probability 变异的概率
N_GENERATIONS = 1000 # 迭代次数DNA_SIZE = len(TARGET_PHRASE) #DNA长度(就是句子长度)
TARGET_ASCII = np.frombuffer(TARGET_PHRASE, dtype=np.uint8) # convert string to number 我们用ASCII代替句子中的字母并表示DNA [64,32,56....]
ASCII_BOUND = [32, 126] #字母范围class GA(object):def __init__(self, DNA_size, DNA_bound, cross_rate, mutation_rate, pop_size):self.DNA_size = DNA_sizeDNA_bound[1] += 1self.DNA_bound = DNA_boundself.cross_rate = cross_rateself.mutate_rate = mutation_rateself.pop_size = pop_size# 随机生成pop_size行,大小为DNA_size的矩阵,即pop_size个DNA序列self.pop = np.random.randint(*DNA_bound, size=(pop_size, DNA_size)).astype(np.int8) # int8 for convert to ASCIIdef translateDNA(self, DNA):"""将DNA翻译成字母:param DNA::return:"""return DNA.tobytes().decode('ascii')def get_fitness(self):"""计算每一个个体的适应度适应度:个体的DNA与TARGET_ASCII有多少个对应位置匹配的个数:return:"""match_count = (self.pop == TARGET_ASCII).sum(axis=1)return match_countdef select(self):"""选择个体成为新的种群可重复选择同一个个体 replace=True:return: 所选个体的下标"""fitness = self.get_fitness() + 1e-4 # 避免适应度为0,不严后面概率为0,就无法选择适应度为0的个体了idx = np.random.choice(np.arange(self.pop_size), size=self.pop_size, replace=True, p=fitness/fitness.sum())return self.pop[idx]def crossover(self, parent, pop):"""交叉配对:param parent::param pop::return:"""if np.random.rand() < self.cross_rate:i_ = np.random.randint(0, self.pop_size, size=1) # select another individual from popcross_points = np.random.randint(0, 2, self.DNA_size).astype(np.bool_) # choose crossover pointsparent[cross_points] = pop[i_, cross_points] # mating and produce one childreturn parentdef mutate(self, child):"""变异:param child::return:"""for point in range(self.DNA_size):if np.random.rand() < self.mutate_rate:child[point] = np.random.randint(*self.DNA_bound) # choose a random ASCII indexreturn childdef evolve(self):pop = self.select()pop_copy = pop.copy()for parent in pop: # for every parentchild = self.crossover(parent, pop_copy)child = self.mutate(child)parent[:] = childself.pop = popif __name__ == '__main__':# 初始化ga = GA(DNA_size=DNA_SIZE, DNA_bound=ASCII_BOUND, cross_rate=CROSS_RATE,mutation_rate=MUTATION_RATE, pop_size=POP_SIZE)# 迭代多次for generation in range(N_GENERATIONS):# 得到每一个个体的适应度fitness = ga.get_fitness()# 打印显示最好的DNAbest_DNA = ga.pop[np.argmax(fitness)]best_phrase = ga.translateDNA(best_DNA)print('Gen', generation, ': ', best_phrase,best_DNA)#得到目标句子就可以结束了if best_phrase == TARGET_PHRASE:breakga.evolve()










-每天了解一点)

— Neck网络介绍(AFPN和BiFPN))




)
——weak_ptr初探)