技能树学徒作业
针对每个癌症的全部基因批量了做了单基因的cox分析,挑选统计学显著的去对应的癌症去打分,看看是否有单细胞亚群特异性。
这题比较常规,但是可以过一遍基础分析的流程。
选择了GSE38832芯片数据用于分析得到cox/logrank显著的基因。GSE200997单细胞数据用于将显著基因映射上去。
芯片数据分析流程-数据下载及提取
1.设置下载时长
rm(list = ls())
#打破下载时间的限制,改前60秒,改后1000w秒
options(timeout = 10000000)
options(scipen = 20)#不要以科学计数法表示
library(tinyarray)
-
传统芯片下载方式
dir.create("./dat")
setwd("./dat")
proj <- "GSE38832"# 下载
geo = geo_download(proj,colon_remove = T)
boxplot(geo$exp[,1:10])
exp = geo$exp
pd = geo$pd
gpl_number = geo$gpl
3.把基因注释好
# 探针注释的获取-----------------
gpl_number <- eSet@annotation;gpl_number
library(tinyarray)
find_anno(gpl_number)
ids <- AnnoProbe::idmap('GPL570')# 加probe_id列,把行名变成一列
library(dplyr)
exp = as.data.frame(exp)
exp = mutate(exp,probe_id = rownames(exp))#加上探针注释
ids = distinct(ids,symbol,.keep_all = T)
exp = inner_join(exp,ids,by ="probe_id")
exp = dplyr::select(exp,-"probe_id")
rownames(exp) <- exp$symbol
exp = dplyr::select(exp,-"symbol")
4.check表达矩阵的数据情况
dim(exp)
range(exp)
boxplot(exp,las = 2) #看是否有异常样本
#数据不齐归一化
exp = limma::normalizeBetweenArrays(exp)
boxplot(exp,las = 2)
-
临床信息和表达矩阵数据对齐
p = identical(rownames(pd),colnames(exp));p
if(!p) {s = intersect(rownames(pd),colnames(exp))exp = exp[,s]pd = pd[s,]
}
6.保存数据
save(pd,exp,gpl_number,proj,file = "step1output.Rdata")
setwd("..")
芯片数据生存分析前的数据整理
1.加载数据和R包
rm(list=ls())
setwd("./dat")
proj <- "GSE38832"
load("step1output.Rdata")library(stringr)
-
临床信息整理
meta <- pd
meta$ID <- rownames(meta)
tmp = data.frame(colnames(meta))
meta = meta[,c('ID','dss_time (disease specific survival time, months)','dss_event (disease specific survival)'
)]
colnames(meta)=c('ID','DSS.time','DSS')
meta$DSS <- gsub("\\(death from cancer\\)|\\(no death\\)","",meta$DSS)
str(meta)
meta$DSS.time <- as.numeric(meta$DSS.time)
meta$DSS <- as.integer(meta$DSS)
3.保存数据
exprSet <- exp
head(rownames(meta))
head(colnames(exprSet))
s = intersect(rownames(meta),colnames(exprSet));length(s)
exprSet = exprSet[,s]
meta = meta[s,]
dim(exprSet)
dim(meta)
identical(rownames(meta),colnames(exprSet))
save(meta,exprSet,proj,file = paste0(proj,"_sur_model.Rdata"))setwd("..")
Cox/Km分析
1.加载数据和R包
rm(list=ls())
setwd("./dat")
proj <- "GSE38832"
load("GSE38832_sur_model.Rdata")
-
批量单因素cox
library(survminer)
library(survival)mySurv <- with(meta, Surv(DSS.time, DSS)) #with函数是一个方便的语法结构,它允许在指定的数据框的上下文中临时评估表达式
cox_results <-apply(exprSet , 1 , function(gene){group=ifelse(gene>median(gene),'high','low') if( length(table(group))<2)return(NULL)survival_dat <- data.frame(group=group,# stage=phe$stage,stringsAsFactors = F)m=coxph(mySurv ~ group, data = survival_dat)beta <- coef(m)se <- sqrt(diag(vcov(m)))HR <- exp(beta)HRse <- HR * se#summary(m)tmp <- round(cbind(coef = beta, se = se, z = beta/se, p = 1 - pchisq((beta/se)^2, 1),HR = HR, HRse = HRse,HRz = (HR - 1) / HRse, HRp = 1 - pchisq(((HR - 1)/HRse)^2, 1),HRCILL = exp(beta - qnorm(.975, 0, 1) * se),HRCIUL = exp(beta + qnorm(.975, 0, 1) * se)), 3)return(tmp['grouplow',])})cox_list <- as.data.frame(t(cox_results))
3.筛选基因并保存数据
# 选择p值小于0.05 和 HR绝对值大于1的基因
cox_deg <- cox_list[(cox_list$p < 0.05) & (abs(cox_list$HR) > 1),]
write.csv(cox_deg, file = "cox_deg.csv")

4.批量log_rank检验
library(survival)
mySurv <- with(meta, Surv(DSS.time, DSS))
logrank_res <- apply(exprSet, 1, function(x){group <- ifelse(x > median(x),"high","low") # 根据表达量的中位数分组fit <- survdiff(mySurv ~ group) #survdiff函数可以分析log_rank检验,核心函数pvalue <- 1-pchisq(fit$chisq, df=1)})res.logrank <- data.frame(symbol = rownames(exprSet), pvalue = logrank_res)
res.logrank <- res.logrank[order(res.logrank$pvalue),]
write.csv(res.logrank, file = "logrank.csv")

4.画一下生存曲线
# 挑选几个基因画
head(cox_list[cox_list[,4]<0.05,])
cg = head(rownames(cox_list[cox_list[,4]<0.05,]))
cg identical(colnames(exprSet), rownames(meta))
mySurv <- with(meta, Surv(DSS.time, DSS))
library(ggstatsplot)
overlap_list <- lapply(cg, function(i){# i = cg[1]survival_dat = metagene = as.numeric(exprSet[i,])survival_dat$gene = ifelse(gene > median(gene),'high','low')table(survival_dat$gene)library(survival)fit <- survfit(Surv(DSS.time, DSS) ~ gene,data = survival_dat)survp = ggsurvplot(fit,data = survival_dat, #这里很关键,不然会报错legend.title = i, #定义图例的名称 # legend = "top",#图例位置# legend.labs = c('High', 'Low'),pval = T, #在图上添加log rank检验的p值# pval.method = TRUE,#添加p值的检验方法risk.table = TRUE, risk.table.y.text = F,xlab = "Time in months", #x轴标题# xlim = c(0, 10), #展示x轴的范围# break.time.by = 1, #x轴间隔size = 1.5, #线条大小ggtheme = theme_ggstatsplot(),palette="nejm", #配色)return(survp)
}) n = length(overlap_list)
n
x=floor(n/2);y=2
# x被定义为n除以2后向下取整的结果
# y被设置为2,图表的布局将有两行
all_plot <- arrange_ggsurvplots(overlap_list,print = F,ncol =x, nrow = y,risk.table.height = 0.3,surv.plot.height = 0.7)
# risk.table.height = 0.3:设置与每个生存曲线图相关联的风险表的高度比例为 0.3
# surv.plot.height = 0.7: 设置生存曲线图本身的高度比例为 0.7。all_plot
ggsave("all_plot.png", plot = all_plot, width = 20, height = 12)setwd("..")

单细胞分析流程
1.数据读取和加载R包
rm(list=ls())
#setwd("./dat/")
library(Seurat)
library(ggplot2)
library(tidyverse)
proj <- "GSE200997"clin <- read.csv("GSE200997_GEO_processed_CRC_10X_cell_annotation.csv.gz",header = T,sep = ",")
sce.raw <- read.csv("GSE200997_GEO_processed_CRC_10X_raw_UMI_count_matrix.csv.gz",header = T,sep = ",")
rownames(sce.raw) <- sce.raw$X
library(dplyr)
sce.raw <- select(sce.raw,-X)
sce <- CreateSeuratObject(sce.raw,project = "GSE200997", min.cells = 3,min.features = 200,)
sce# 把临床信息辅助到seurat对象中
phe <- sce@meta.data
# 首先check一下官方提供的GEO文件中的行顺序和sce对象中的细胞顺序是否一致
identical(rownames(phe),clin$X)
#[1] TRUEphe <- cbind(phe,clin)
phe$samples <- sub("B","N",phe$samples)
phe <- select(phe, -X)
sce@meta.data <- phesave(sce,file = "sce.raw.Rdata")
2.质量控制
Idents(sce) <- sce$samples
# 质量控制-看线粒体基因,核糖体基因,红细胞
#计算线粒体,核糖体和血红蛋白基因比例并添加到数据中
sce[["percent.mt"]] <- PercentageFeatureSet(sce, pattern = "^MT-")
sce[["percent.rp"]] <- PercentageFeatureSet(sce, pattern = "^RP[SL]") #核糖体
sce[["percent.hb"]] <- PercentageFeatureSet(sce, pattern = "^HB[^(P)]")VlnPlot(sce, features = c("nFeature_RNA","nCount_RNA", "percent.mt","percent.rp","percent.hb"), ncol = 3,pt.size = 0, group.by = "samples")#散点图-----Count_RNA和线粒体
plot1 <- FeatureScatter(sce, feature1 = "nCount_RNA", feature2 = "percent.mt")
#散点图-----Count_RNA和细胞数?
plot2 <- FeatureScatter(sce, feature1 = "nCount_RNA", feature2 = "nFeature_RNA")
#散点图-----Count_RNA和核糖体
plot3 <- FeatureScatter(sce, feature1 = "nCount_RNA", feature2 = "percent.rp")
#散点图-----核糖体和线粒体
plot4 <- FeatureScatter(sce, feature1 = "percent.rp", feature2 = "percent.mt")
plot1 + plot2 + plot3 + plot4# 设置质控的标准
sce <- subset(sce,percent.mt < 25 & #也可以更加严格小于5nFeature_RNA > 200 & nFeature_RNA < 6000 #& #nfeature值可以严格到<2500#nCount_RNA < 18000 &#percent.rp <15 &#percent.hb <1
)
4.对数据标准化及显示高变基因
# 标准化数据
sce <- NormalizeData(sce, normalization.method = "LogNormalize", scale.factor = 10000)
# 鉴定2000个高变基因(数量可人为设置,一般是2K)
sce <- FindVariableFeatures(sce, selection.method = "vst", nfeatures = 2000)
# Identify the 10 most highly variable genes
top10 <- head(VariableFeatures(sce), 10)
# 对高变基因可视化
# 对高可变基因进行可视化
plot1 <- VariableFeaturePlot(sce)
plot2 <- LabelPoints(plot = plot1, points = top10, repel = TRUE)
plot1 + plot2 #这个图片窗口需要拉大一点
5.缩放数据
# In addition we scale the data(缩放数据),保证每个基因在同一个尺度上
all.genes <- rownames(sce)
sce <- ScaleData(sce, features = all.genes)
# 这个ScaleData 函数的 Default is variable features.
# 如果我们不添加 features = all.genes ,
# 它就是默认的前面的 FindVariableFeatures 函数的2000个基因
-
PCA降维
# PCA降维
sce <- RunPCA(sce, features = VariableFeatures(object = sce) , #npcs = 20,verbose = FALSE)
sce@reductions$pca@cell.embeddings[1:4,1:4]
# 展示1和2主成分中的主要“荷载”
VizDimLoadings(sce, dims = 1:2, reduction = "pca")
# Dimplot可视化
DimPlot(sce, reduction = "pca")
#heatmap可视化(可自行修改dims)
DimHeatmap(sce, dims = 1, cells = 500, balanced = TRUE)#接下来我们既然已经对庞大的数据进行了降维(也就是聚堆)的形式,那么我们究竟要选择几个PC来代表这么庞大的数据呢,肯定不能都选,否则我的数据量还是这些,就没有我们前面一直渗透的缩小缩小再缩小的含义了
sce <- JackStraw(sce, num.replicate = 100)
sce <- ScoreJackStraw(sce, dims = 1:20) #dims最大是20
#上面两个代码是通过不同的方式来帮助我们选择PC的数目,并且分别都对应不同的可视化
JackStrawPlot(sce, dims = 1:20) #最大值是20
ElbowPlot(sce,ndims=50)
#不同的判断方法选择的PC数是不一样的,而且理论上来说保存更多的PC可以保存更多的生物学差异,所以这里我们灵活选择即可,因为都不算错
7.cluster-分群-PCA可视化
# 基于降维后的数据构建细胞邻近图
# dims的数量是根据上一步所确定的ElbowPlot
dims_N <- "30"
sce <- FindNeighbors(sce, dims = 1:dims_N, verbose = FALSE) # 进行聚类分析,基于邻近图(之前由FindNeighbors 函数构建)划分细胞群体
# 可以通过Idents(sce) 对比前后的levels数量变化
# 请注意,我这边resolution设置了一个梯度
sce <- FindClusters(sce, resolution = seq(0.1, 1.2, by = 0.1), verbose = FALSE)
head(Idents(sce), 5)
#resolution大小决定cluster的数量(0.4-1.2范围)#可视化cluster
DimPlot(sce, reduction = "pca")
8.clustree图
library(clustree)
library(patchwork)
p1 <- clustree(sce, prefix = 'RNA_snn_res.') + coord_flip()
#这里的RNA_snn_res后面的数值是可以修改的
p2 <- DimPlot(sce, group.by = 'RNA_snn_res.0.5', label = T)
p1 + p2 + plot_layout(widths = c(3, 1))
-
UMAP/tSNE可视化
#UMAP/tSNE可视化前先确定一个想要的reslution值
#这里重新跑一遍之后后面就会按照新跑的reslution值进行分析
sce <- FindClusters(sce, resolution = 0.5, verbose = FALSE)
head(Idents(sce), 5)#假设电脑的显卡非常高级的话,可以不用PCA降维,直接UMAP
#Umap方式
sce <- RunUMAP(sce, dims = 1:dims_N, umap.method = "uwot", metric = "cosine")
DimPlot(sce,label = T)
#Tsne方式
#pbmc <- RunTSNE(pbmc )
#DimPlot(pbmc,label = T,reduction = 'tsne',pt.size =1)#数据保存
phe=sce@meta.data
save(phe,file = 'phe-by-basic-seurat.Rdata')#把seurat得到的meta.data信息保存下来
10.细胞亚群注释前marker提取
#首先要确认一下每一cluster中的marker基因
#其中pct.1:在目标细胞簇中表达的细胞比例;pct.2:在其他细胞簇中表达的细胞比例。
sce.markers <- FindAllMarkers(sce, only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25, verbose = FALSE)
head(sce.markers,n = 5)
#显示每个簇中log2FC表达最高5个值
a <- sce.markers %>%group_by(cluster) %>% # dplyr包中的函数可以进行分组操作top_n(n = 5, wt = avg_log2FC)%>% # dplyr包中的函数,可以提取前多少行data.frame()#可视化
DimPlot(sce, reduction = "umap", group.by = 'seurat_clusters',label = TRUE, pt.size = 0.5) #请注意下边的feature参数中的基因是要存在于pbmc.markers中
#feature参数不能重复
DotPlot(sce, features = c("CD45", "PTPRC", #immune"EPCAM",#epithelial"CD10","MME","CD31","PECAM1" #stromal),group.by = 'seurat_clusters') + theme(axis.text.x=element_text(angle=45,hjust = 1))
11.cluster合并
#####细胞生物学命名
#先创建一个表格
celltype=data.frame(ClusterID=0:23,celltype= 0:23)
celltype[celltype$ClusterID %in% c(0:4,6,8,12,16,18,20,23 ),2]='immune'
celltype[celltype$ClusterID %in% c(5,7,9,11,13:15,17,21),2]='epithelial'
celltype[celltype$ClusterID %in% c(10,19),2]='stromal'
celltype[celltype$ClusterID %in% c(22),2]='Unknow'sce@meta.data$celltype = "NA" #先在scRNA中添加celltype的一行
for(i in 1:nrow(celltype)){sce@meta.data[which(sce@meta.data$RNA_snn_res.0.5 == celltype$ClusterID[i]),'celltype'] <- celltype$celltype[i]}
table(sce@meta.data$celltype)# 修改Idents中分群编号为细胞类型
Idents(sce) <- sce$celltype
DimPlot(sce, reduction = "umap", label = TRUE, repel = T,pt.size = 0.5)
scCustomize::DimPlot_scCustom(sce,figure_plot = TRUE)step1.final = sce
save(step1.final,proj,file = 'step1.final.Rdata')
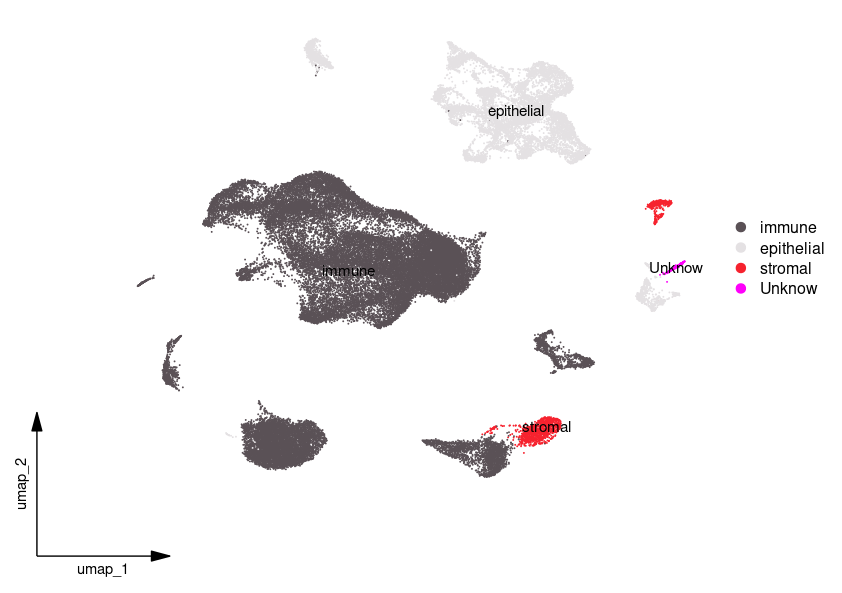
对细胞亚群的命名比较粗糙,其中不能确定的群我设置为了unknow群
获得研究者提供的Epi基因然后对自己数据进行基因划分
1.导入数据加载R包
rm(list = ls())
library(Seurat)
library(openxlsx)
load("step1.final.Rdata")
sce <- step1.final
cox_deg <- read.csv("cox_deg.csv",header = T, sep = ",")
logRank <- read.csv("logrank.csv",header = T,sep = ",")
Author_define <- read.xlsx("41586_2022_5402_MOESM4_ESM (1).xlsx")
EpiGenes <- Author_define$gene[1:99]
2.cox_deg 交集
Epi_cox <- intersect(cox_deg$X,EpiGenes)
# 排除在 Epi_cox 中的基因
remaining_genes <- setdiff(cox_deg$X, Epi_cox)
Immue_cox <- cox_deg$X[cox_deg$X %in% remaining_genes]#表达量的另一种绘图方式
library(Nebulosa)
plot_density(sce,features = Epi_cox) +plot_layout(ncol = 2)
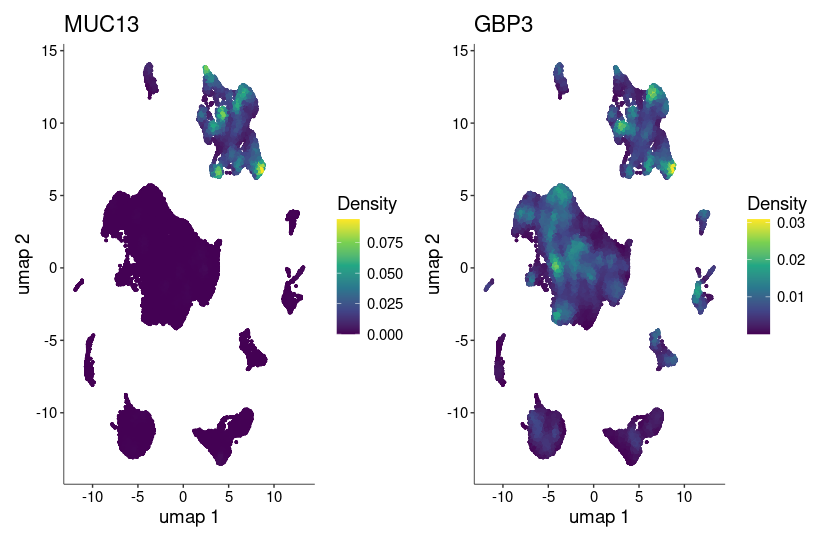
这里只交集到了两个基因,所以绘制了表达量图看一下
Addmodulescore评分-All
sce = AddModuleScore(object = sce,features = list(cox_deg$X))
colnames(sce@meta.data)
p = FeaturePlot(sce,'Cluster1') #默认名称cluster1
p # 常规绘图
dat<- data.frame(sce@meta.data, sce@reductions$umap@cell.embeddings,seurat_annotation = sce@active.ident)
class_avg <- dat %>%group_by(celltype) %>% #按照seurat_annotation列(即细胞的分类)对数据进行分组。summarise(umap_1 = median(umap_1),umap_2 = median(umap_2) #对每个分组计算UMAP坐标的中位数 画label)library(ggpubr)
ggplot(dat, aes(umap_1, umap_2)) +geom_point(aes(colour = Cluster1)) + #修改这里的colourviridis::scale_color_viridis(option="A") +ggrepel::geom_label_repel(aes(label = celltype),data = class_avg,label.size = 0,segment.color = NA)+theme_bw()
ggsave(filename="addmodulescore_all.png",width = 9,height = 7)

cox显著的基因在几个亚群中均有表达,其中在immune细胞群中似乎表达最高
Addmodulescore评分-EPI
sce = AddModuleScore(object = sce,features = list(Epi_cox))
colnames(sce@meta.data)
p = FeaturePlot(sce,'Cluster1') #默认名称cluster1
p # 常规绘图
dat<- data.frame(sce@meta.data, sce@reductions$umap@cell.embeddings,seurat_annotation = sce@active.ident)
class_avg <- dat %>%group_by(celltype) %>% #按照seurat_annotation列(即细胞的分类)对数据进行分组。summarise(umap_1 = median(umap_1),umap_2 = median(umap_2) #对每个分组计算UMAP坐标的中位数 画label)library(ggpubr)
ggplot(dat, aes(umap_1, umap_2)) +geom_point(aes(colour = Cluster1)) + #修改这里的colourviridis::scale_color_viridis(option="A") +ggrepel::geom_label_repel(aes(label = celltype),data = class_avg,label.size = 0,segment.color = NA)+theme_bw()
ggsave(filename="addmodulescore.png",width = 9,height = 7)
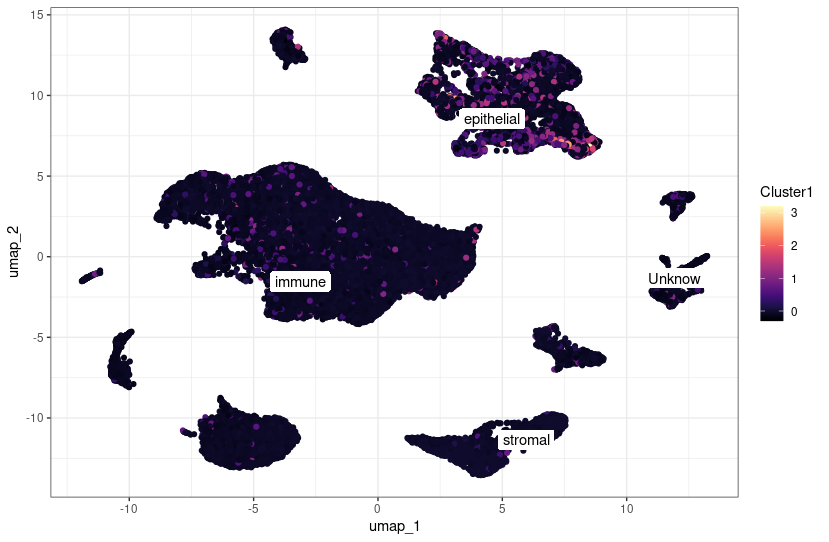
Epi相关的基因就集中在epthelial
logrank显著的基因就不做了
致谢:感谢曾老师,小洁老师以及生信技能树团队全体成员(部分代码来源:生信技能树马拉松和数据挖掘课程)。
注:若对内容有疑惑或者有发现明确错误的朋友,请联系后台(希望多多交流)。更多内容可关注公众号:生信方舟
- END -




)






)
)

)


![SpringBoot学习06-[SpringBoot与AOP、SpringBoot自定义starter]](http://pic.xiahunao.cn/SpringBoot学习06-[SpringBoot与AOP、SpringBoot自定义starter])

