新手遇到的问题都是类似的,比如批量ID转换

虽然我写过大量的教程:ID转换大全 不过都需要R基础,因为是大批量转换啊!
但热心肠的植物生物信息学教学大佬还是友善的给出了解决方案
我也狗尾续貂制作了一个网页工具教程:
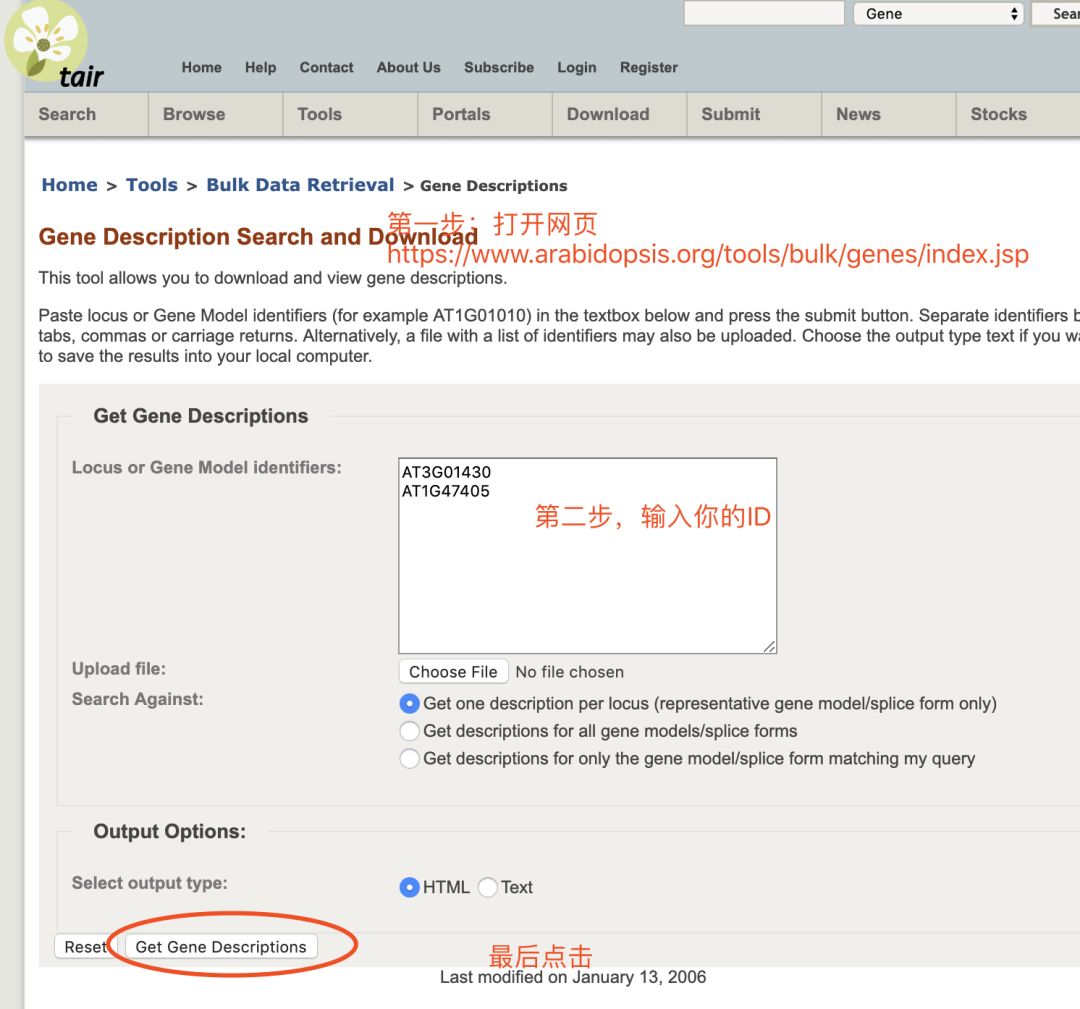
简单的3个步骤,不会代码也可以很容易把ID批量转换啦!
不过有趣的是我搜索电脑资料,看到了2年前写的拟南芥教程。
不过我为什么会花时间写拟南芥教程呢?

1 首先加载必要的包
library(ggplot2)
library(stringr)
# source("https://bioconductor.org/biocLite.R")
# biocLite("clusterProfiler")
# biocLite("org.At.tair.db")
library(clusterProfiler)
library(org.At.tair.db)
2 然后导入数据
deg1=read.table('https://raw.githubusercontent.com/jmzeng1314/my-R/master/DEG_scripts/tair/DESeq2_DEG.Day1-Day0.txt')
deg1=na.omit(deg1)
head(deg1)
## baseMean log2FoldChange lfcSE stat pvalue
## AT3G01430 22.908929 18.989990 2.148261 8.839704 9.597263e-19
## AT1G47405 20.709551 20.958806 2.434574 8.608820 7.381677e-18
## AT2G33830 1938.159722 -2.560609 0.312663 -8.189678 2.619266e-16
## AT5G28627 8.118376 -21.131318 2.875691 -7.348257 2.008078e-13
## AT2G33750 9.789740 -19.989301 2.847513 -7.019915 2.220033e-12
## AT3G54500 2238.314652 2.720430 0.386305 7.042182 1.892517e-12
## padj
## AT3G01430 1.858318e-14
## AT1G47405 7.146571e-14
## AT2G33830 1.690562e-12
## AT5G28627 9.720602e-10
## AT2G33750 7.164418e-09
## AT3G54500 7.164418e-09
prefix='Day1-Day0'
3 然后判断显著差异基因
很明显,这个时候用padj来挑选差异基因即可,不需要看foldchange了。
table(deg1$padj<0.05)
##
## FALSE TRUE
## 19166 197
table(deg1$padj<0.01)
##
## FALSE TRUE
## 19303 60
diff_geneList = rownames(deg1[deg1$padj<0.05,])
up_geneList = rownames(deg1[deg1$padj<0.05 & deg1$log2FoldChange >0,])
down_geneList = rownames(deg1[deg1$padj<0.05 & deg1$log2FoldChange <0,])
length(diff_geneList)
## [1] 197
length(up_geneList)
## [1] 89
length(down_geneList)
## [1] 108
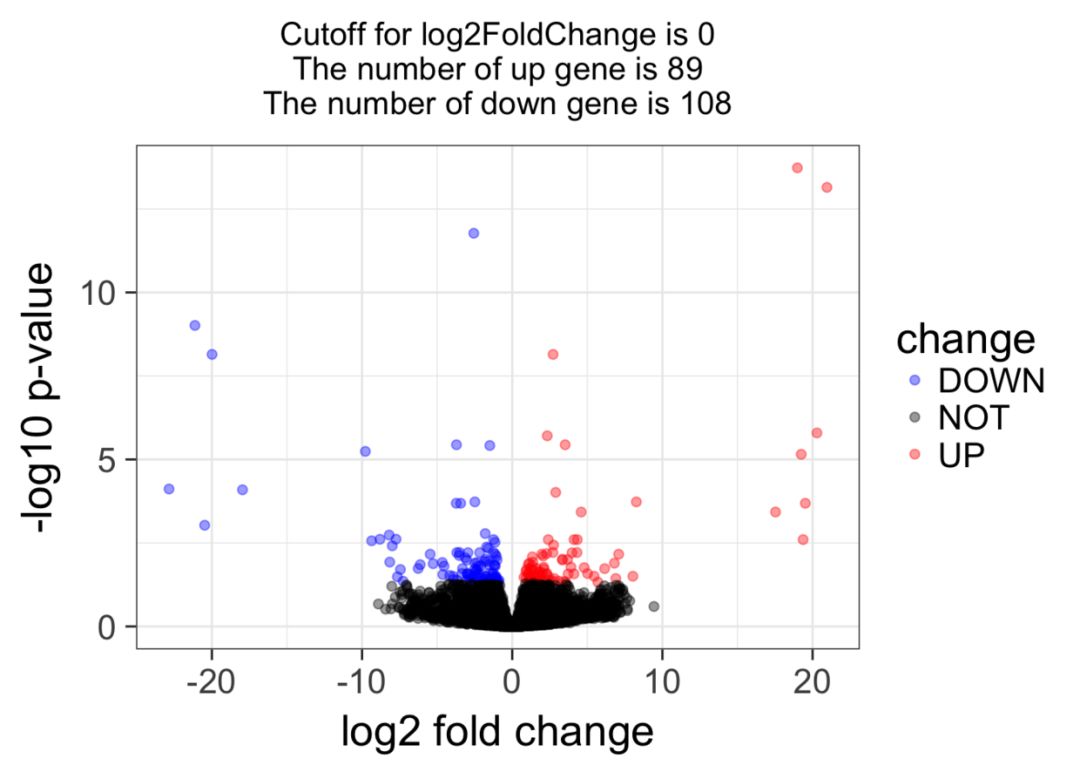
3.1 画个火山图看看挑选的差异基因合理与否
colnames(deg1)
## [1] "baseMean" "log2FoldChange" "lfcSE" "stat"
## [5] "pvalue" "padj"
log2FoldChange_Cutof = 0
## 这里我不准备用log2FoldChange来挑选差异基因,仅仅是用padj即可
deg1$change = as.factor(ifelse(deg1$padj 0.05 &
abs(deg1$log2FoldChange) > log2FoldChange_Cutof,
ifelse(deg1$log2FoldChange > log2FoldChange_Cutof ,'UP','DOWN'),'NOT'))
this_tile 'Cutoff for log2FoldChange is ',round(log2FoldChange_Cutof,3),
'\nThe number of up gene is ',nrow(deg1[deg1$change =='UP',]) ,
'\nThe number of down gene is ',nrow(deg1[deg1$change =='DOWN',])
)
g_volcano = ggplot(data=deg1, aes(x=log2FoldChange, y=-log10(padj), color=change)) +
geom_point(alpha=0.4, size=1.75) +
theme_set(theme_set(theme_bw(base_size=20)))+
xlab("log2 fold change") + ylab("-log10 p-value") +
ggtitle( this_tile ) +
theme(plot.title = element_text(size=15,hjust = 0.5))+
scale_colour_manual(values = c('blue','black','red')) ## corresponding to the levels(res$change)
print(g_volcano)
4 GO/KEGG注释
4.1 首先进行KEGG注释
diff.kk <- enrichKEGG(gene = diff_geneList,organism = 'ath',pvalueCutoff = 0.99,qvalueCutoff=0.99
)kegg_diff_dt <- as.data.frame(setReadable(diff.kk,org.At.tair.db,keytype = 'TAIR'))up.kk <- enrichKEGG(gene = up_geneList,organism = 'ath',pvalueCutoff = 0.99,qvalueCutoff=0.99
)kegg_up_dt <- as.data.frame(setReadable(up.kk,org.At.tair.db,keytype = 'TAIR'))down.kk <- enrichKEGG(gene = down_geneList,organism = 'ath',pvalueCutoff = 0.99,qvalueCutoff=0.99
)kegg_down_dt <- as.data.frame(setReadable(down.kk,org.At.tair.db,keytype = 'TAIR'))4.2 可视化看看KEGG注释结果
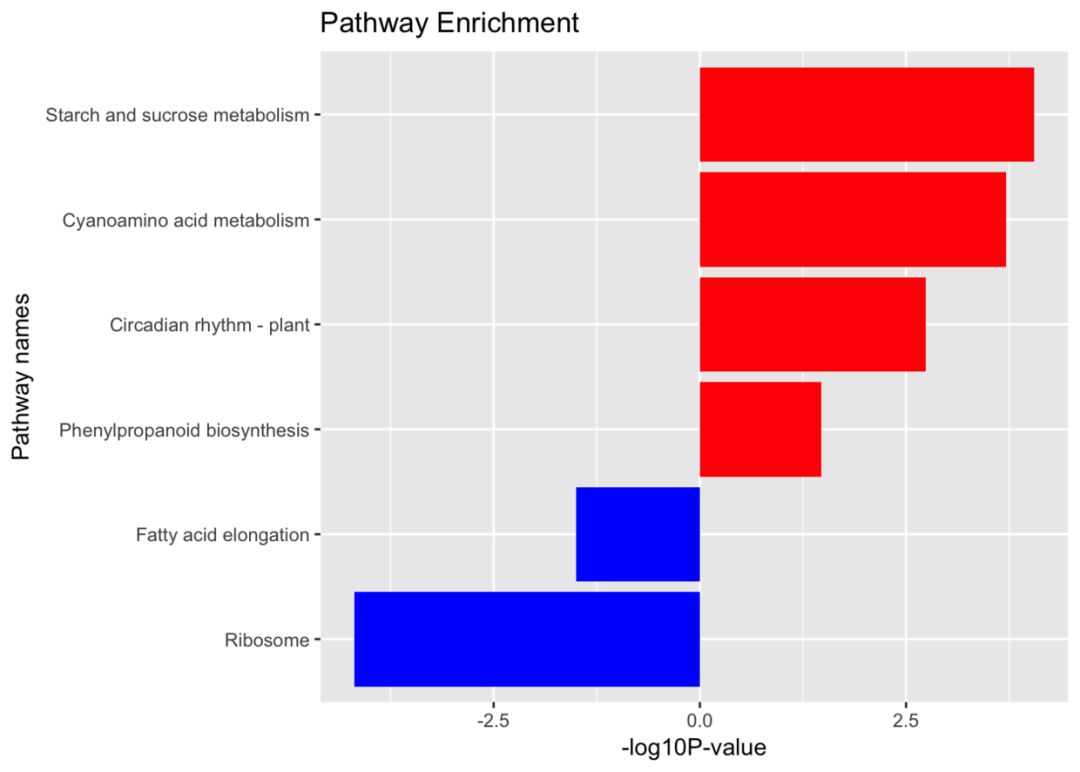
## KEGG patheay visulization:
down_kegg$pvalue<0.05,];down_kegg$group=-1
up_kegg$pvalue<0.05,];up_kegg$group=1
dat=rbind(up_kegg,down_kegg)
colnames(dat)
## [1] "ID" "Description" "GeneRatio" "BgRatio" "pvalue"
## [6] "p.adjust" "qvalue" "geneID" "Count" "group"
dat$pvalue = -log10(dat$pvalue)
dat$pvalue=dat$pvalue*dat$group
dat=dat[order(dat$pvalue,decreasing = F),]
g_kegg geom_bar(stat="identity") +
scale_fill_gradient(low="blue",high="red",guide = FALSE) +
scale_x_discrete(name ="Pathway names") +
scale_y_continuous(name ="-log10P-value") +
coord_flip() +
ggtitle("Pathway Enrichment")
print(g_kegg)
4.3 接着进行GO注释
for (onto in c('CC','BP','MF')){
ego OrgDb = org.At.tair.db,
keyType = 'TAIR',
ont = onto ,
pAdjustMethod = "BH",
pvalueCutoff = 0.2,
qvalueCutoff = 0.9)
ego 'TAIR')
write.csv(as.data.frame(ego),paste0(prefix,"_",onto,".csv"))
#ego2
ego2=ego
pdf(paste0(prefix,"_",onto,'_barplot.pdf'))
p=barplot(ego2, showCategory=12)+scale_x_discrete(labels=function(x) str_wrap(x,width=20))print(p)dev.off()
}ggsave(filename = paste0(prefix,"_volcano_plot.pdf"),g_volcano)## Saving 7 x 5 in image
ggsave(filename = paste0(prefix,"_kegg_plot.pdf"),g_kegg)
## Saving 7 x 5 in image
write.csv(x = kegg_diff_dt,file = paste0(prefix,"_kegg_diff.csv"))
write.csv(x = kegg_up_dt,file = paste0(prefix,"_kegg_up.csv"))
write.csv(x = kegg_down_dt,file = paste0(prefix,"_kegg_down.csv"))
5 基因ID注释
library(org.At.tair.db)
ls('package:org.At.tair.db')
## [1] "org.At.tair" "org.At.tair.db"
## [3] "org.At.tairARACYC" "org.At.tairARACYCENZYME"
## [5] "org.At.tairCHR" "org.At.tairCHRLENGTHS"
## [7] "org.At.tairCHRLOC" "org.At.tairCHRLOCEND"
## [9] "org.At.tairENTREZID" "org.At.tairENZYME"
## [11] "org.At.tairENZYME2TAIR" "org.At.tairGENENAME"
## [13] "org.At.tairGO" "org.At.tairGO2ALLTAIRS"
## [15] "org.At.tairGO2TAIR" "org.At.tairMAPCOUNTS"
## [17] "org.At.tairORGANISM" "org.At.tairPATH"
## [19] "org.At.tairPATH2TAIR" "org.At.tairPMID"
## [21] "org.At.tairPMID2TAIR" "org.At.tairREFSEQ"
## [23] "org.At.tairREFSEQ2TAIR" "org.At.tairSYMBOL"
## [25] "org.At.tair_dbInfo" "org.At.tair_dbconn"
## [27] "org.At.tair_dbfile" "org.At.tair_dbschema"
## Then draw GO/kegg figures:
deg1$gene_id=rownames(deg1)
id2symbol=toTable(org.At.tairSYMBOL)
deg1=merge(deg1,id2symbol,by='gene_id')
## 可以看到有一些ID是没有gene symbol的,这样的基因,意义可能不大,也有可能是大部分人漏掉了
head(deg1)
## gene_id baseMean log2FoldChange lfcSE stat pvalue
## 1 AT1G01010 58.25657 1.13105335 0.8000274 1.4137683 0.1574300
## 2 AT1G01010 58.25657 1.13105335 0.8000274 1.4137683 0.1574300
## 3 AT1G01020 542.64394 -0.05745554 0.3721650 -0.1543819 0.8773086
## 4 AT1G01030 48.77442 -1.09845263 1.2875862 -0.8531100 0.3935983
## 5 AT1G01040 1708.68949 0.32421734 0.2777530 1.1672865 0.2430947
## 6 AT1G01040 1708.68949 0.32421734 0.2777530 1.1672865 0.2430947
## padj change symbol
## 1 0.6008903 NOT ANAC001
## 2 0.6008903 NOT NAC001
## 3 0.9805661 NOT ARV1
## 4 0.8144858 NOT NGA3
## 5 0.6992279 NOT DCL1
## 6 0.6992279 NOT EMB60
可以看到基因的ID和symbol的对应关系就出来了,根使用网页工具是类似的,感兴趣的朋友可以试试看网页工具和R代码的ID批量转换差别有多大。
■ ■ ■
全国巡讲约你
第1-10站北上广深杭,西安,郑州, 吉林,武汉和成都(全部结束)
七月份我们不外出,只专注单细胞!
系统学习单细胞分析,报名生信技能树的线下培训,手慢无 。
。
一年一度的生信技能树单细胞线下培训班火热招生
全国巡讲第11站-港珠澳专场(生信技能树爆款入门课)












)
的功能是:将长整型数中每一位上为奇数的数依次取出,构成一个新数放在冲。 - 赏学吧...)





